Table 2. Sensitivity analysis for human ES and GM12878 cells (replicate 1 data from ENCODE Broad database) percentages using various methods.
| unit-mean | quantile | MACS | ChIPDiff | rank | RSEG | two-stage unit-mean | ChIPnorm | |
| thresholds |
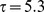
|

|
p-val 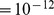
|
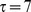
|
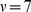
|
cdf 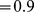
|
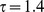
|
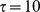
|
| GM12878 differential (four-fold) expressed: 1927 genes | ||||||||
| sensitivity (HES K27-enriched) | 11.26 | 11.26 | 11.26 | 10.85 | 17.85 | 8.27 | 10.69 | 11.05 |
| error (GM12878 K27-enriched) | 0.10 | 0.21 | 0 | 0.16 | 5.35 | 0 | 0.31 | 0.16 |
| HES differential (four-fold) expressed: 2908 genes | ||||||||
| sensitivity (GM12878 K27-enriched) | 1.55 | 10.97 | 1.79 | 4.09 | 33.29 | 0.06 | 14.65 | 12.00 |
| error (HES K27-enriched) | 1.38 | 0.93 | 3.54 | 1.20 | 2.27 | 11.81 | 1.20 | 0.55 |
Experiments: unit-mean; quantile; MACS peak finder; ChIPDiff; Rank normalization; two-stage unit-mean; ChIPnorm. The parameters of all the methods (except rank normalization) were adjusted so that all of them give almost the same percentage (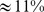 ) of experiment “sensitivity (HES K27-enriched)”.
) of experiment “sensitivity (HES K27-enriched)”.
