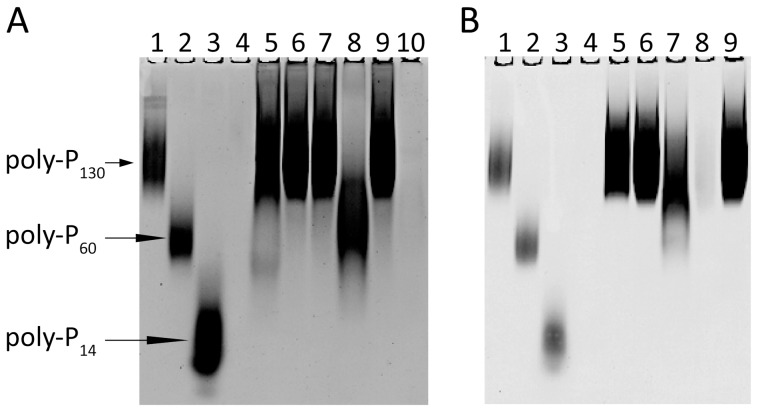
Figure 2. Hydrolysis of polyphosphate (poly-P130) mediated by cell free extracts of Escherichia coli CF6032 (ΔgppA Δppkx) and Mycobacterium smegmatis mc2155, supplemented by MTB-PPX1, Rv1026 or E. coli PPX proteins.
Crude cell lysates were prepared from of cultures of E. coli MG1655 (wild type), E. coli CF6032 (triple mutant defective for GPP, PPX and PPK expression) and M. smegmatis mc2155 (see materials and methods). Cell lysate (2 µg total protein), polyphosphate substrate (poly-P130, 0.1 mM) and the MTB-PPX1, Rv1026, E. coli PPX (EC-PPX), or maltose binding protein (MBP, negative control) proteins (2 µg) were incubated at 37°C for 2 hours in 50 mM HEPES pH 6.8, 25 mM KCl, 1 mM MnCl2 (100 µl). Reaction mixtures were resolved on TBE 12% polyacrylamide gels, and stained with toluidine blue, to analyze the polyphosphate products formed. Poly-P60 and poly-P14 (RegeneTiss) were included as polyphosphate chain length standards. Panel A Lane 1: poly-P130; lane 2: poly-P60; lane 3: poly-P14; lane 4: empty; lane 5: E. coli MG1655 lysate + poly-P130; lane 6: E. coli CF6032 lysate + poly-P130; lane 7: CF6032 + poly-P130 + MBP; lane 8: CF6032 + poly-P130 + MTB-PPX1, lane 9: CF6032 + poly-P130 + Rv1026; lane 10: CF6032+ poly-P130 + E. coli PPX (EC-PPX). Panel B. Lane 1: poly-P130 ; lane 2: poly-P60; lane 3: poly-P14; lane 4: empty; lane 5: M. smegamtis mc2155 lysate + poly-P130; lane 6: mc2155 + poly-P130 + MBP; lane 7: mc2155 + poly-P130 + MTB-PPX1, lane 8: mc2155 + poly-P130 + EC-PPX; lane 9: mc2155 + poly-P130 + Rv1026.