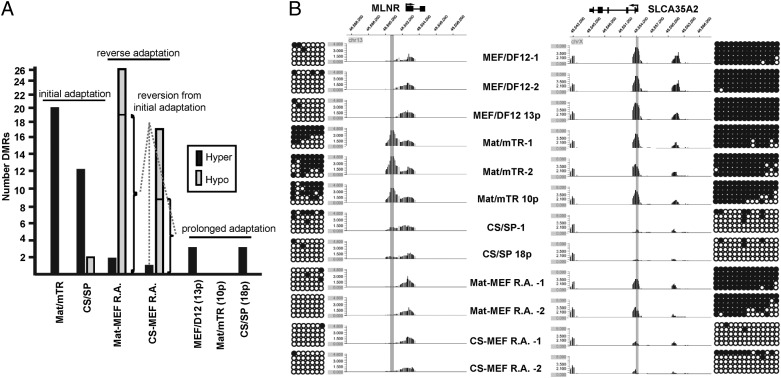
Fig. 3.
DNA methylation in hESCs during culture adaptation. (A) Summary of DMRs across samples. Mat/mTR and CS/SP initial DMRs were identified through comparison with original MEF/DF12-cultured cells. Prolonged DMRs were identified in higher passages versus initial adaptation to Mat/mTR or CS/SP, or in the case of MEF/DF12, p82 versus start culture (p69). Sample descriptions provide passage number postadaptation. Reversible DMRs are noted. Hyper = hypermethylation; Hypo = hypomethylation. (B) Examples of and validation of culture-specific DMRs. A hypermethylated Mat/mTR-specific DMR is shown on the left at the MLNR gene promoter. The peak was identified in three separate Mat/mTR samples and is reversible in accordance with culture conditions. A hypomethylated CS/SP DMR at the SLCA35A2 promoter region is provided on the right. The DMR is an example of a rare irreversible methylation change. MIRA-chip results were validated by bisulfite sequencing of sequences centric to identified DMRs (highlighted in gray). Circles represent consecutive CpG dinucleotides. Dark circles: methylated CpG sites. Open circles: unmethylated CpG sites.