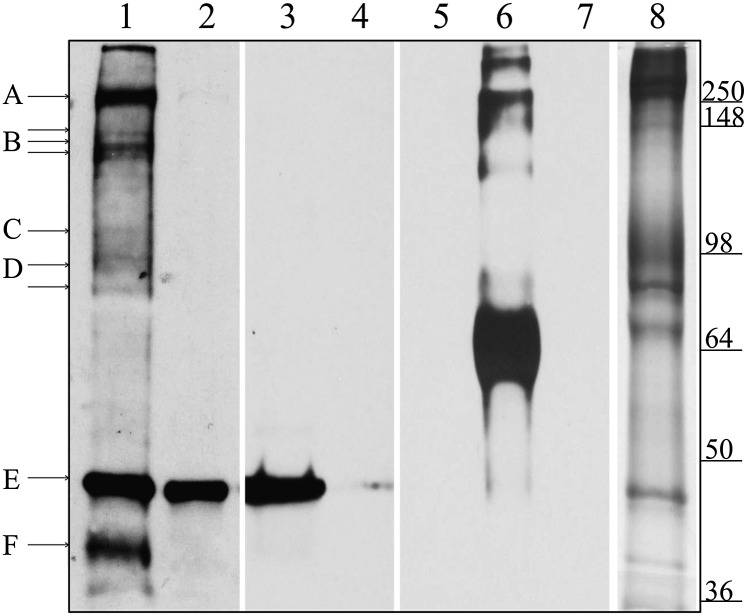
Fig. 3.
Identification of ghost proteins labeled with pool-associated N3-ATP. Here the ghosts were filled or emptied of N3-ATP, as described in Materials and Methods. After washing, the membranes were photolyzed and separated by SDS/PAGE. Equal amounts of ghost proteins (30 μg) were loaded in all lanes. Lanes 1–7 were transferred to nitrocellulose and immunoblotted with an antibody to N3-ATP, whereas lane 8 was stained with Coomassie blue. Lane 1 shows pools filled with N3-ATP; lanes 2 and 3, pools first filled with N3-ATP and then emptied by either running the Na+ pump forward or running PGK reaction backward, respectively. In lane 4, pools are filled with N3-ATP in the presence of excess unlabeled ATP to block any specific ATP-binding sites. Lanes 5–7 show the specificity of the N3-ATP antibody, demonstrating that it labels only BSA photolyzed with N3-ATP (lane 6), not unlabeled BSA (lane 5) or unlabeled RBC ghosts (lane 7). Lane 8 shows a Coomassie blue stain of ghost proteins after separation by SDS/PAGE. The proteins tentatively identified are indicated with lettered lines. Possible protein candidates for the labeled bands are A, β-spectrin and/or ankyrin; B, Ca2+ pump (Mr ∼140 kDa) or fragments of spectrin/ankyrin; C, α-subunit of the Na+/K+ pump (Mr ∼110 kDa); D, band 3; E, actin; and F, GAPDH.