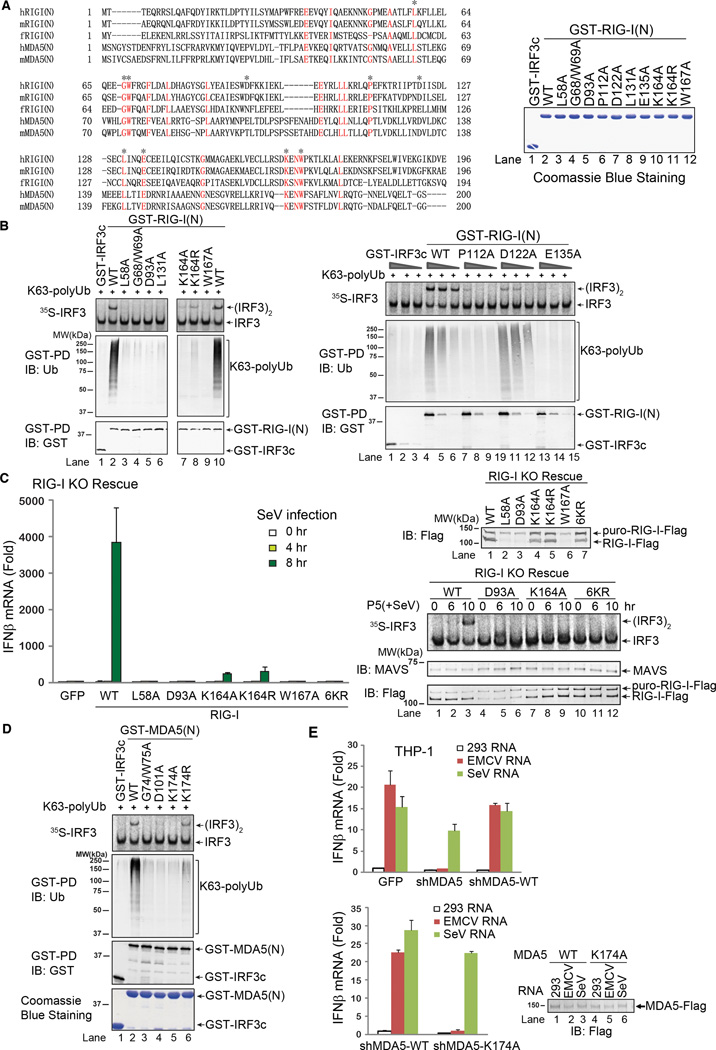
Figure 3.
Ubiquitin binding is essential for RIG-I and MDA5 activation. A. Sequence alignment of the N-termini of RIG-I and MDA5 from human (h) and mouse (m) as well as zebrafish (f) RIG-I N-terminus. Asterisks indicate the residues to be mutated in this study. Shown on the right are the GST-RIG-I(N) mutant proteins expressed in and purified from E. coli. B. Point mutants of RIG-I(N) were incubated with K63 polyUb and then analyzed by GST pull-down and IRF3 dimerization assays. C. Full-length RIG-I WT and mutants were stably expressed in RIG-I knockout (KO) MEF cells. The reconstituted cells were infected with SeV for the indicated time, and then IFNβ was measured by qPCR (left). Mitochondrial fractions (P5) were prepared from the virus-infected cells, and MAVS activity was measured in IRF3 dimerization assay (bottom right). The RIG-I proteins in the reconstituted cells were immunoblotted with a RIG-I antibody (upper right). The upper band denotes a fusion protein in which RIG-I was not cleaved from the puromycin resistance gene product due to incomplete self-cleavage of the 2A peptide in the lentiviral vector. D. Point mutants of MDA5(N) were tested for K63 polyUb binding and IRF3 activation as in B. E. THP-1 cells stably expressing a lentiviral shRNA vector targeting MDA5 or those in which endogenous MDA5 was replaced with WT or K174A MDA5 were transfected with indicated RNA, and then IFNβ induction was measured by qPCR. Results shown are representatives of two experiments.