Abstract
Selective inhibition of disease-related proteins underpins the majority of successful drug–target interactions. However, development of effective antagonists is often hampered by targets that are not druggable using conventional approaches. Here, we apply engineered zinc-finger protein transcription factors (ZFP TFs) to the endogenous phospholamban (PLN) gene, which encodes a well validated but recalcitrant drug target in heart failure. We show that potent repression of PLN expression can be achieved with specificity that approaches single-gene regulation. Moreover, ZFP-driven repression of PLN increases calcium reuptake kinetics and improves contractile function of cardiac muscle both in vitro and in an animal model of heart failure. These results support the development of the PLN repressor as therapy for heart failure, and provide evidence that delivery of engineered ZFP TFs to native organs can drive therapeutically relevant levels of gene repression in vivo. Given the adaptability of designed ZFPs for binding diverse DNA sequences and the ubiquity of potential targets (promoter proximal DNA), our findings suggest that engineered ZFP repressors represent a powerful tool for the therapeutic inhibition of disease-related genes, therefore, offering the potential for therapeutic intervention in heart failure and other poorly treated human diseases.
Introduction
Effective human therapies typically require an agent that blocks function of a pathological gene product. Development of such agents can be problematic if the drug target—which is typically a protein—is located in an inaccessible cellular compartment, or presents a mechanism of action that is not readily disrupted. Addressing the gap between known and druggable disease targets requires a methodology that can circumvent such limitations; this consideration has kindled great interest in methods capable of decreasing the cellular levels of these targeted proteins by suppressing their expression (e.g., the use of RNA interference technology to suppress the translation of targeted mRNAs). It has been demonstrated that human genes implicated in disease may be regulated by targeting genomic DNA using designed zinc-finger transcription factors (ZFP TFs) composed of an engineered zinc-finger DNA-binding domain specific for the gene of interest fused to a relevant functional domain (reviewed in ref. 1). We and others have previously shown that designed ZFP TFs can evoke therapeutic activation of gene expression in vivo.2,3,4,5 ZFP repressors have also been reported to function in a tumor xenograft model and in mouse retina against a human transgene.6,7 However, as a class of therapeutic molecules that have the potential to provide antagonist activity to the large body of nondruggable targets, ZFP repressors have not been tested extensively in vivo, especially in the context of bona fide endogenous genes and delivery to native organs/tissues.
Phospholamban (PLN), a 52 amino acid protein, plays a critical role in the kinetics of calcium flux in cardiac muscle. It inhibits the activity of the sacroplasmic reticulum calcium ATPase (SERCA2a),8 which returns cytosolic calcium into sacroplasmic reticulum after each contractile cycle in preparation for subsequent excitation-contraction events.9 Decreased SERCA2a activity, either as a result of reduced expression, increased PLN expression, or PLN mutations that superinhibit SERCA2a function has been associated with human heart failure.10,11,12,13 Conversely, increasing the SERCA2/PLN ratio in cardiomyocytes, either by reducing PLN levels or increasing SERCA2 levels, markedly improves both the systolic contraction and diastolic relaxation of cardiac muscle.14,15,16,17,18 Moreover, dominant-negative PLN strategies that interfere with the PLN–SERCA2 interaction have proven effective in augmenting SERCA2 function and cardiac contractility in animal models.19 Importantly, unlike β-agonists, direct targeting of SERCA2 or PLN enhances contractility without concomitant activation of the entire β-adrenergic pathway, which has been shown to increase mortality in congestive heart failure patients. While inhibition of PLN provides an attractive approach for improving cardiac function, its mechanism of action (as an inhibitor of SERCA2a) and location within an organelle membrane combine to make it a difficult target for standard small molecule or antibody-based methods.20 We sought to determine whether engineered ZFP TFs can be applied to the PLN gene to achieve therapeutically relevant gene repression and enhancement of cardiac function.
Results
Design and in vitro testing of ZFP TFs
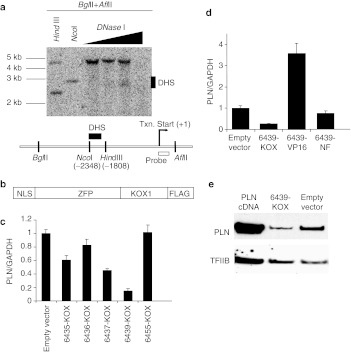
Rat H9C2(2-1) cells are derived from heart tissues and express detectable levels of PLN, therefore were chosen as a cell culture model for identifying ZFP repressors of PLN. ZFP designs were focused on a region of accessible chromatin as revealed by mapping of DNaseI hypersensitive sequences. In H9C2(2-1) cells, a single accessible region was identified between bases –2,350 and –2,050 relative to the transcription start site (Figure 1a). This region included a span of enhanced sequence conservation between rat and human, suggesting regulatory significance. ZFP TFs were engineered to bind five distinct 18-bp targets within this region. Each ZFP comprised six fingers, and was constructed from an archive of 2-finger units as previously described.21 Such ZFPs have demonstrated a capacity for potent target repression in vitro with single-gene specificity in expression profiling studies.22 Each was then linked to the Krüppel-associated box (KRAB A/B) repression domain from the N-terminal region of the KOX1 protein to create a ZFP TF repressor (Figure 1b). Transient transfection of plasmid DNAs encoding these ZFP TFs into H9C2(2-1) cells demonstrated up to an 80% reduction in PLN mRNA, with the protein 6439-KOX reproducibly exhibiting the greatest degree of repression (Figure 1c). Further studies of this protein indicated that its repressive effect was dependent upon the functional domain since removal of the KOX moiety eliminated repression and replacement with the VP16 activation domain increased PLN mRNA levels (Figure 1d). Western blot analysis confirmed that 6439-KOX also reduced PLN protein levels (Figure 1e). Together, these results demonstrate that 6439-KOX is a potent and domain-dependent repressor of PLN.
Figure 1.
Targeting and in vitro analysis of zinc-finger protein transcription factor (ZFP TF) repressors of the rat phospholamban (PLN) gene. (a) Nuclei of H9C2(2-1) cells were isolated and titrated with DNaseI (Worthington) as described.3 Isolated genomic DNA was double-digested with BglII and AflII. Aliquots of double-digested DNA were subjected to HindIII or NcoI digest for use as genomic size markers. The location of the DNaseI hypersensitive region (DHS) is indicated by a solid bar next to the blot and in the schematic diagram. The transcription start site (+1) is marked by the hooked arrow. The nucleotide locations of the NcoI site and the HindIII site are relative to the transcription start site. The open box indicates the location of the probe for Southern blot. (b) The structure of the assembled ZFP repressors is depicted in the diagram. NLS, nuclear localization signal; KOX1, repression domain of KOX1; FLAG, FLAG epitope tag. (c) H9C2(2-1) cells were transiently transfected with plasmids expressing the indicated ZFP TFs (Supplementary Materials and Methods). After 72 hours mRNA was harvested and the levels of PLN mRNA and GAPDH mRNA were quantified using real-time reverse transcription (RT)-PCR (Taqman). The PLN level was normalized to that of GAPDH. (d) Variants of the 6439-KOX repressor protein in which the KOX repression domain was either removed (6439-NF) or replaced with an activator (6439-VP16), were tested for function in H9C2(2-1) cells. PLN mRNA levels were measured as in b. (e) H9C2(2-1) cells were transiently transfected as indicated, and 72 hours later whole cell extracts were collected and subjected to western blot analysis. A PLN cDNA was used as a positive control for the anti-PLN antibody.
ZFP-driven PLN repression is highly specific
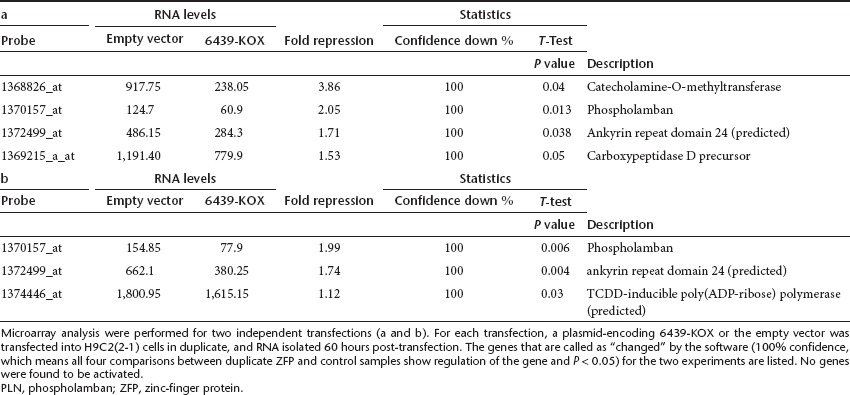
6439-KOX recognizes an 18-bp sequence that is unique within the rat genome, theoretically permitting function with singular specificity. We conducted DNA microarray experiments to determine the genome-wide impact of 6439-KOX on gene expression. The genes regulated by 6439-KOX (as determined by the software) in two independent transfections are listed in Table 1a and b. No gene was found to be activated. Only two genes, PLN and ankyrin repeat domain 24 (predicted), were found to be repressed in both experiments. The regulation of PLN (2.05-fold and 1.99-fold) is more pronounced than that of the other gene (1.71 fold and 1.74 fold). The smaller fold of repression for PLN as measured by microarray (relative to the approximately fivefold-repression measured by real-time reverse transcription-PCR (Supplementary Figure S1) likely reflects differences in how RNA levels are measured in these two distinct assays. Specifically, the fold of repression may be underestimated in the microarray experiment because the PLN levels (120–155 fluorescence units in control samples and 61–78 units in ZFP-treated samples, as measured by the scanner) is near the lower end of the signal range genome-wide (10–25,000 fluorescence units). The repression of catecholamine-O-methyltransferase observed in the first microarray experiments could not be confirmed by Taqman analysis (Supplementary Figure S1), therefore is likely a result of aberrant detection of the microarray. These data demonstrate that the engineered TF 6439-KOX functions with high specificity within the monitored genome, with PLN potentially being the only gene that is significantly regulated.
Table 1. Specificity of the ZFP repressor of PLN.
Designed PLN repressor accelerates calcium transients and improves contractility
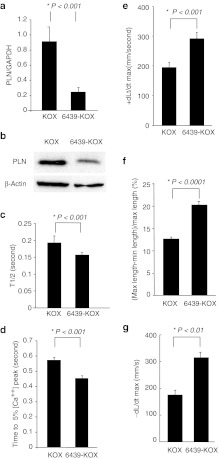
As a more rigorous and therapeutically relevant test of function, we examined the activity of 6439-KOX in primary rat cardiomyocytes. Cardiomyocytes are the target cell type for treating heart failure and express much higher levels of PLN than H9C2(2-1) cells. Accordingly, primary cardiomyocytes isolated from day 1 neonatal rats were transduced with recombinant adenovirus encoding either 6439-KOX (Ad-6439-KOX) or the KOX repression domain alone (Ad-KOX). Under conditions yielding >85% transduction, Ad-6439-KOX repressed PLN mRNA levels by up to 75% relative to Ad-KOX (Figure 2a). Western blot analysis confirmed a significant reduction in PLN protein levels by 6439-KOX (Figure 2b), demonstrating potent inhibition of the PLN promoter even in its highly induced native state in cardiomyocytes.
Figure 2.
Zinc-finger protein (ZFP)-driven repression of phospholamban (PLN) improves calcium transient and contractility in rat cardiomyocytes. (a,b) Cultured neonatal rat cardiomyocytes were transduced with adenoviral vectors encoding either the PLN repressor ZFP (6439-KOX) or the repressor domain alone without the ZFP DNA-binding domain (KOX) at MOI 200. Seventy-two hours later, PLN (a) mRNA levels and (b) protein levels were measured. (c,d) Cultured neonatal rat cardiomyocytes infected with either Ad-6439-KOX or Ad-KOX were loaded with Fluo-3 and calcium transients were assessed during excitation-contraction induced by electrical pacing. The rate of calcium reuptake into the sacroplasmic reticulum was measured as the time required to decrease cytosolic calcium levels to (c) 50% or (d) 5% of their peak after depolarization. (n = 25 for KOX-transduced cells, n = 25 for 6439-KOX-transduced cells). (e,f) Either Ad-6439-KOX or Ad-KOX, mixed with an equal titer of Ad-FS-Red (an adenoviral vector encoding a Fluorescent marker) were codelivered to rat hearts via direct intramyocardial injections. Cardiomyocytes were isolated 72 hours after injection, and edge-detection-based analysis of single-cell contractility during electrical stimulation was performed on FS-Red-positive cells. The rate of (e) cell lengthening (relaxation), (f) percent cell shortening and the (g) rate of cell shortening (contraction) were determined (n = 15 cells per group three separate injected hearts per treatment; five cotransduced cells per heart examined; and at least five separate contraction-relaxation cycles were analyzed per cell).
To determine whether this level of PLN repression would be sufficient to elicit the anticipated effects on calcium handling, neonatal rat cardiomyocytes were transduced with either Ad-6439-KOX or Ad-KOX, and 48 hours later calcium transients were measured in electrically paced cardiomyocytes loaded with the calcium fluorophore Fluo-3. The times needed to reduce the cytosolic calcium levels to 50% of the peak levels (T1/2) (Figure 2c) and to 5% of the peak levels (Figure 2d) were significantly decreased in Ad-6439-KOX-treated cells relative to control cells, indicating an increased rate of calcium removal from the cytosol during each pacing cycle that is consistent with reduced PLN levels and enhanced SERCA2a activity.
To analyze the effects of 6439-KOX on contractility at the single-cell level we carried out an in vivo direct intramyocardial coinjection of either Ad-6439-KOX and Ad-FS-Red (an adenovirus encoding a fluorescent marker, FS-Red) or Ad-KOX and Ad-FS-Red. We and others have previously observed that when delivering two distinct adenoviral vectors simultaneously a high-cotransduction rate occurs (Supplementary Figure S2).23 Thus, FS-Red labeled cells are likely to be also successfully transduced with the test adenoviral vectors. At 72 hours postinjection, hearts were explanted and cardiomyocytes isolated, and the contractility of individual FS-Red-positive cardiomyocytes was determined by an edge detection method during electrical pacing.24 As shown in Figure 2e, cardiomyocytes transduced with Ad-6439-KOX exhibited a highly significant enhancement in the maximum rate of lengthening when compared to cells isolated following injection of the Ad-KOX vector, consistent with the expected increase in SERCA2a activity in these cells. Moreover, both the extent of cell shortening (Figure 2f) as well as the rates of shortening (Figure 2g) were also significantly improved in cells transduced with Ad-6439-KOX. These data demonstrate that at the individual cardiomyocyte level, ZFP-mediated PLN repression is sufficient to drive a phenotype of enhanced calcium transient and contractility.
ZFP-driven PLN repression improves heart function, alters the hypertrophic response, and reduces post-infarction cardiac dysfunction
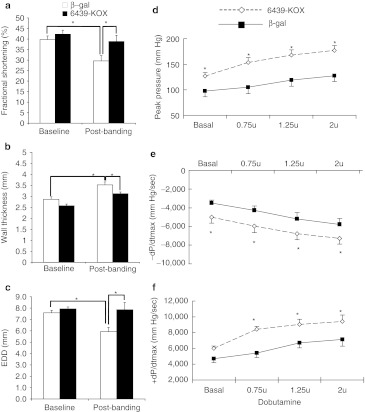
To test the in vivo efficacy of the PLN repressor we used a rat model of sequential cardiac pressure overload and myocardial infarction, allowing us to assess the effects of PLN repression on two commonly encountered clinical pathophysiological states. Pressure overload was induced by ligature-induced stenosis of the abdominal aorta between the renal arteries. The degree of stenosis was standardized surgically and corroborated by Doppler flow analysis across the stenotic region. We used this model as it induces not only pressure overload, but also alterations in the renin–angiotensin system that have been associated with heart failure.25 Before aortic constriction we performed gene delivery to the heart by direct multisite intramyocardial injection of adeno-associated virus serotype 2 vectors (AAV-2) expressing either 6439-KOX or β-galactosidase (as control). Intramyocardial injection of AAV-β-gal in this manner resulted in expression primarily restricted to portions of the anterior and lateral walls of the left ventricle (Supplementary Figure S3). Evaluation by echocardiography 4.5 weeks later revealed that the rats injected with AAV-6439-KOX had augmented cardiac contractile function (as measured by fractional shortening; Figure 3a), and an attenuated hypertrophic response (less thickening of the ventricle wall, Figure 3b). These observations are consistent with the predicted phenotype of PLN repression and increased SERCA2a activity which include reduced hypertrophy.9,26 Interestingly, whereas end-diastolic diameters were unchanged from baseline in the 6439-KOX-treated hearts at 4.5 weeks post-aortic banding, they were reduced significantly in the AAV-β-gal hearts that received aortic banding (Figure 3c), possibly secondary to a greater hypertrophic response in these hearts.
Figure 3.
An engineered zinc-finger protein transcription factor (ZFP TF) repressor improves cardiac function in vivo in a rat model of heart failure. (a–c) AAV-2 vectors encoding either the phospholamban (PLN) repressor 6439-KOX or β-gal control were delivered to the hearts of juvenile rats by direct intramyocardial injection. Immediately after gene transfer the rats underwent induction of pressure overload by intrarenal constriction of the abdominal aorta. Four and half-weeks later echocardiography analysis was used to determine (a) percent fractional shortening, (b) ventricular wall thickness, and (c) end-diastolic diameters (EDD); baseline: before aortic banding; post-banding: 4.5 weeks after aortic banding. (e,f) Four and half-weeks after gene transfer and induction of pressure overload, myocardial infarction was induced by ligation of the left anterior descending coronary artery. Fourteen-days later terminal hemodynamic analysis was carried out to determine (d) peak developed pressures, (e) the rate of relaxation (–dP/dt), and (f) the rate of systolic pressure generation (+dP/dt), both at baseline and during infusion of the inotropic drug dobutamine at graduated doses. (n = 11 for the 6439-KOX group; n = 10 for the β-gal group; *P value <0.05).
As a further test of 6439-KOX's ability to preserve cardiac function after a pathophysiological insult, we induced myocardial infarction in these same rats 4.5 weeks after aortic constriction. Infarction was induced by ligation of the left anterior descending coronary artery, the major blood supply of the anterior wall of the left ventricle. Fourteen-days later, terminal hemodynamic studies with concomitant pharmacological stress (dobutamine, a β-adrenergic agonist) revealed significantly greater peak developed ventricular pressures (Figure 3d) and higher rates of intraventricular pressure change during both relaxation and contraction in animals injected with AAV-ZFP6439-KOX (–dP/dt and +dP/dt, respectively; Figure 3e, f), although heart rates were not different between the groups. Together these data confirm that ZFP TF repression of the endogenous PLN gene results in the expected alterations in cardiac contractility and function.
Discussion
We report the development of a highly specific ZFP TF engineered to bind a DNaseI-accessible region of the rat PLN promoter. This ZFP TF potently represses PLN transcription in multiple cell types and functionally reduces PLN protein levels as gauged by western blot analysis as well as measurements of Ca2+ transients and contractility in ZFP-treated rat cardiomyocytes. DNA microarray analysis reveals that the specificity of the PLN repressor approaches the level of single-gene regulation within the monitored genome. Moreover, the ZFP repressor is active in vivo: in a rat model of heart failure, ZFP delivery improves several measures of cardiac performance that are consistent with functional repression of PLN. These data thus provide an important proof-of-concept that ZFP-mediated gene repression may be employed in vivo for the potential treatment of heart failure.
The cardiac benefits of increased calcium flux into the sarcoplasmic reticulum are well-established,20 and modulation of PLN and/or SERCA2a activity levels has been highlighted as an attractive strategy for improving calcium handling with minimal collateral effects. This recognition has driven interest in diverse approaches for increasing the SERCA2/PLN ratio, including cardiac delivery of SERCA2a cDNA, a dominant-negative PLN variant as well as RNAi against PLN, which all have been shown to improve contractile properties and prevent the progression of heart failure in animal models.14,19,27 Indeed, a SERCA2a gene therapy approach has been shown to be safe and improved cardiac function in early and mid-stage human clinical trials for congestive heart failure.28,29 The ZFP repressor that we have developed provides a new approach for increasing SERCA2a activity and improving calcium handling, therefore potentially expanding our options for this serious disease.
A particularly encouraging result in the current work relates to the potency of the effects observed using the PLN-specific ZFP repressor despite the relatively modest efficiency of myocardial delivery (Supplementary Figure S3). Even such restricted delivery of the ZFP repressor to the anterior and lateral walls of the left ventricle led to improved measures of cardiac function. While more efficient delivery to a much larger proportion of the myocardium is likely to be required for maximum efficacy, especially in human subjects, delivery methods that improve the transduction efficiency of the myocardium (e.g., catheter-based antegrade or retrograde intracoronary myocardial gene transfer30,31,32), as well as alternative AAV serotypes that improve transduction efficiency in heart33 have been described for this purpose.
Beyond repressing PLN expression, our study provides direct in vivo evidence that “subtractive” therapy, by far the most common mode of action for existing drugs, can be achieved against a recalcitrant drug target (through targeting its genomic DNA) by an engineered ZFP repressor. This result has significant implications for our ability to therapeutically modulate otherwise refractory drug targets in the heart and other organs and tissues. For many such targets (exemplified by PLN), subcellular location (e.g.,, within organellar membranes) or mechanism-of-action (e.g., as a ligand) render the protein product resistant to inhibition via small molecule or antibody-based methods. For other recalcitrant targets, the presence of dozens if not hundreds of other highly similar family members (e.g., kinases, ion channels) can confound attempts to develop inhibitors with sufficient selectivity for therapeutic use. Engineered ZFP repressors circumvent such limitations since the diverse nature of small molecule drug–target interactions can be reduced to a well-understood array of protein-DNA contacts for any drug target. Moreover, due to the higher degree of genetic drift that occurs within regulatory (noncoding) regions of the genome compared to that of coding regions, even genes encoding highly related protein family members are likely to possess unique DNA sequences in their promoters that can be targeted with high specificity by engineered ZFPs.
While engineered ZFPs regulate gene expression by exploiting the natural mechanism of transcription, RNAi-based approaches provide a different way of inhibiting difficult drug targets by triggering the degradation of the mRNA encoding the gene product. For phenotypic penetrance RNAi may need to address the many hundreds (potentially thousands) of copies of the mRNA expressed from the target gene. In contrast, the ZFP TF approach addresses the source of this message—namely the DNA template, which is present at two copies per cell (for autosomal genes). Moreover, by targeting 18-bp sites and using simple bioinformatics approaches to avoid targeting repetitive sequences, ZFP-driven gene regulation can be highly specific. Because specificity data was not provided for siRNA against PLN,27 we cannot directly compare that to the specificity of the ZFP. Nevertheless, this report adds to a growing list of ZFPs developed by our group and others that demonstrate exquisite specificity,5,22,34 which compares favorably to that reported for RNAi-based methods.35,36,37 Notably a recent study of PLN shRNA in healthy canines revealed cardiac toxicity that the investigators attribute to shRNA-induced perturbation of miRNA expression rather than downregulation of PLN activity.38 ZFP-driven PLN repression as a direct transcriptional repressor offers an alternative strategy for PLN inhibition that does not engage the RNAi processing machinery. This approach thus warrants testing for safety and efficacy in larger animals.
Finally, in striving to extend our therapeutic options to include the genome as a bona fide drug target, the versatility of DNA recognition permitted by designed transcription factors is paramount. The C2H2 class of ZFPs possess a combination of modularity and adaptability that is unmatched in nature.39,40 During the past decade we, and others, have engineered ZFPs for specific targeting of numerous genes (reviewed in refs. 1,41,42). Given this versatility and the therapeutic benefits of targeting genomic DNA, designed ZFPs have the potential of becoming a new class of drugs capable of potent and selective modulation of virtually any therapeutic gene target.
Materials and Methods
Mapping of DNaseI-accessible regions. Nuclei isolation from H9C2(2-1) cells, DNaseI treatment, genomic DNA isolation, restriction enzyme digestion and Southern blot analysis were performed as described.3
ZFP design and construction. To target the DNaseI hypersensitive region of the rat PLN gene, six-finger ZFP TFs were assembled as described.21,43 The resultant ZFP gene was cloned into the pcDNA3.1 vector (Invitrogen, Grand Island, NY ) as a fusion with the KRAB A/B transcriptional repression domain of KOX1 as previously described.22 The amino acid residues of the helical regions of ZFP-6439 responsible for specific DNA binding (Fingers 1–6) are TSADLTE, ASANLSR, RSDALST, DRSTRTK, RSDVLSA, and DRSNRIK. The binding site of ZFP-6439 is: gacatggccatggatagc.
Generation of viral vectors. Recombinant adenoviral vectors encoding 6439-Kox and Kox were generated using the Adeno-X Express System (Clontech, Mountain View, CA). Adenoviral particle numbers were determined by absorbance at 260 nm and infectious titers were determined using the Adeno-X Rapid Titer Kit (Clontech).
Recombinant adeno-associated vectors encoding 6439-kox and β-gal were generated using the triple transfection method as described.44
Cell culture and transient transfection. H9C2(2-1) cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Duplicate transfections were performed for each construct using FuGene 6 (Promega, Madison, WI). Cells were harvested 48–72 hours post-transfection for RNA isolation or making whole cell extract. For Affymetrix analysis, ~1 × 106 cells H9C2(2-1) cells were transfected in duplicate with 7 µg of ZFP6439-KOX or pcDNA3 empty vector. Cells were harvested 60 hours after transfection. Neonatal and adult rat cardiac myocytes were prepared as previously described.18,24
Real-time reverse transcription-PCR analysis. Real-time reverse transcription-PCR was performed as previously described.5 The levels of PLN mRNA and GAPDH mRNA were quantified using standard curves spanning a 125-fold concentration range. Each RNA sample was assayed in duplicate Taqman reactions. The ratio of PLN/GAPDH was used to determine the normalized levels of PLN.
Western blot analysis. Seventy-two hours after transfection of H9C2(2-1) cells or infection of rat cardiomyocytes, whole cell extracts were analyzed by western blot as previously described22 using a monoclonal anti-PLN antibody (Upstate Biotech, Billerica, MA). A polyclonal anti-TFIIB or anti-β-actin antibody (Santa Cruz Biotech, Santa Cruz, CA) was used for internal loading control.
Affymetrix analysis. Total RNA was isolated from transfected H9C2(2-1) cells using TRIzol reagent (Invitrogen) according to manufacturer's recommendations. RNA samples for hybridization were prepared according to standard Affymetrix protocol using 10 µg of total RNA. Changes in gene expression were analyzed using rat RAE-230A array, which contains 15,923 probes representing roughly 14,000 genes. Data analysis was carried out using Affymetrix Microarray Suite 5.0 and Data Mining Tool 3.0 software. Criteria for differentially expressed genes were: 100% confidence call, and P value <0.05.
Single-cell Ca2+ transient analysis. Neonatal cardiomyocytes from Sprague Dawley rats were infected with Ad-6439-KOX or Ad-KOX at MOI 200 in a Petri dish invested with platinum electrodes and placed on the heated stage of a Zeiss Axiovert 135 inverted microscope fitted with a Delta Scan Photometry system (Photon Technology International, Lawrenceville, NJ) for measurement of calcium transients. The cells were paced at a rate of 1 Hz. Fluo-3 (Molecular Probes, San Diego, CA) was loaded into myocytes according to the manufacturer's recommendations and was used as an indicator of cytosolic calcium. Calcium transients were captured using Felix software (Photon Technology International) and background subtracted. The time for intracellular calcium to decrease to 50% of maximum amplitude (T1/2) was determined as described.18,24
Single-cell contractility analysis. Sprague Dawley rats were treated humanely in accordance with federal and Yale School of Medicine policies regarding the use of animals in research. Ad-6439-KOX or Ad-KOX (2.5 × 109 pfu) were coinjected along with an adenovirus encoding a fluorescent marker, Ad-FS-Red (2.5 × 109 pfu), directly into the myocardium of rats via a lateral thoracotomy as previously described.45 Seventy-two hours later rod-shaped cardiac myocytes were isolated from these hearts by collagenase digestion as described.24 Isolated cardiac myocytes were paced at 1 Hz, and a video edge detection system (Crescent Electronics, Windsor, Ontario, Canada) measured the magnitude of cell shortening. The percentage of cell shortening was expressed as ([basal length-contracted length]/basal length) × 100. Rates of contraction and relaxation were calculated as basal length—shortest length/time from basal to shortest length (+dP/dt); and shortest length—basal length/time from shortest to basal length during relaxation –dP/dt).
In vivo transduction with AAV vectors. Myocardial gene delivery was accomplished by direct intramyocardial injection with a modified 30 gauge bore needle. Briefly, in anesthetized and ventilated rats the heart was exposed by lateral thoracotomy. The modified needle was fashioned with a near 90 angle at the distal tip to facilitate entry into the myocardium through the thoracotomy. AAV-β-gal or AAV-6439-KOX (5 × 1011 AAV vector genomes) was injected at six separate sites in a total of 200 µl of phosphate-buffered saline (volume equally divided amongst the sites). After injection the chest was closed in layers, and air evacuated from the pleural space via a cannula left in place during chest closure.
Induction of pressure overload, myocardial infarction, echocardiography, and hemodynamic analysis. All in vivo experiments were performed according protocols approved by the IACUC at Yale Medical School. Abdominal aortic banding was accomplished by a transabdominal surgical approach. The infrarenal abdominal aorta was visualized and a silk ligature placed around it and an 18-gauge needle angled at the tip. After ligation the needled was removed and the abdominal closed in layers. The degree of stenosis and documentation of flow to the aorta beyond the constriction was determined by ultrasound and Doppler imaging. Transthoracic echocardiography was performed 4.5 weeks later under inhaled anesthesia (isofluorane) using a 7.5 MHz transducer.24,46 Following echocardiography myocardial infarction was induced by lateral thoracotomy followed by ligation of the left anterior descending coronary artery just after the first branch. Ligation and cessation of myocardial flow in that territory was confirmed by visualization of myocardial blanching. The chest was then closed in layers and the animals allowed to recover. Fourteen-days after infarction cardiac function was assessed by terminal hemodynamic studies in ventilated animals under isofluorane anesthesia, using a microtransducer-tipped catheter inserted into the left ventricle via the carotid artery. Graduated doses of dobutamine were infused at a constant rate via a syringe pump and a cannula in the internal jugular vein.46
SUPPLEMENTARY MATERIAL Figure S1. Catecholamine-O-methyltransferase expression is not regulated by the PLN repressor. Figure S2. Efficiency of coinfection following intramuscular injection of two separate adenovirus vectors. Figure S3. AAV-mediated delivery following direct intramyocardial injection. Supplementary Materials and Methods.
Acknowledgments
We thank Ken Kim, Kathie Howes, and Simon Chandler for ZFP assembly; Albert Sinusas for assistance with the rat model, Fyodor Urnov, Susan Abrahamson, and Lei Zhang for helpful discussions and comments, and Edward Lanphier for encouragement and support. This study is supported in part by NIH NHLBI grants HL64001 and HL63770 to F.J.G. H.S.Z., D.G., M.K., S.H., Y.L., L.H., S.K.S., C.C.C., E.J.R., and P.D.G. are employees of Sangamo BioSciences Inc.
Supplementary Material
Catecholamine-O-methyltransferase expression is not regulated by the PLN repressor.
Efficiency of coinfection following intramuscular injection of two separate adenovirus vectors.
AAV-mediated delivery following direct intramyocardial injection.
REFERENCES
- Klug A. Towards therapeutic applications of engineered zinc finger proteins. FEBS Lett. 2005;579:892–894. doi: 10.1016/j.febslet.2004.10.104. [DOI] [PubMed] [Google Scholar]
- Lu Y, Tian C, Danialou G, Gilbert R, Petrof BJ, Karpati G.et al. (2008Targeting artificial transcription factors to the utrophin A promoter: effects on dystrophic pathology and muscle function J Biol Chem 28334720–34727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebar EJ, Huang Y, Hickey R, Nath AK, Meoli D, Nath S.et al. (2002Induction of angiogenesis in a mouse model using engineered transcription factors Nat Med 81427–1432. [DOI] [PubMed] [Google Scholar]
- Laganiere J, Kells AP, Lai JT, Guschin D, Paschon DE, Meng X.et al. (2010An engineered zinc finger protein activator of the endogenous glial cell line-derived neurotrophic factor gene provides functional neuroprotection in a rat model of Parkinson's disease J Neurosci 3016469–16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi K, Zhang HS, Kachi S, Balaggan KS, Yu Q, Guschin D.et al. (2007Gene transfer of an engineered zinc finger protein enhances the anti-angiogenic defense system Mol Ther 151917–1923. [DOI] [PubMed] [Google Scholar]
- Kang YA, Shin HC, Yoo JY, Kim JH, Kim JS., and, Yun CO. Novel cancer antiangiotherapy using the VEGF promoter-targeted artificial zinc-finger protein and oncolytic adenovirus. Mol Ther. 2008;16:1033–1040. doi: 10.1038/mt.2008.63. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Sanges D, Marrocco E, Bonetti C, Di Vicino U, Marigo V.et al. (2011Zinc-finger-based transcriptional repression of rhodopsin in a model of dominant retinitis pigmentosa EMBO Mol Med 3118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan DH., and, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- Frank KF, Bölck B, Erdmann E., and, Schwinger RH. Sarcoplasmic reticulum Ca2+-ATPase modulates cardiac contraction and relaxation. Cardiovasc Res. 2003;57:20–27. doi: 10.1016/s0008-6363(02)00694-6. [DOI] [PubMed] [Google Scholar]
- Haghighi K, Chen G, Sato Y, Fan GC, He S, Kolokathis F.et al. (2008A human phospholamban promoter polymorphism in dilated cardiomyopathy alters transcriptional regulation by glucocorticoids Hum Mutat 29640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi K, Kolokathis F, Gramolini AO, Waggoner JR, Pater L, Lynch RA.et al. (2006A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy Proc Natl Acad Sci USA 1031388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamisawa S, Sato Y, Tatsuguchi Y, Fujino T, Imamura S, Uetsuka Y.et al. (2003Mutation of the phospholamban promoter associated with hypertrophic cardiomyopathy Biochem Biophys Res Commun 3041–4. [DOI] [PubMed] [Google Scholar]
- Nef HM, Möllmann H, Skwara W, Bölck B, Schwinger RH, Hamm Ch.et al. (2006Reduced sarcoplasmic reticulum Ca2+ -ATPase activity and dephosphorylated phospholamban contribute to contractile dysfunction in human hibernating myocardium Mol Cell Biochem 28253–63. [DOI] [PubMed] [Google Scholar]
- del Monte F, Williams E, Lebeche D, Schmidt U, Rosenzweig A, Gwathmey JK.et al. (2001Improvement in survival and cardiac metabolism after gene transfer of sarcoplasmic reticulum Ca(2+)-ATPase in a rat model of heart failure Circulation 1041424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar RJ, Schmidt U, Matsui T, Guerrero JL, Lee KH, Gwathmey JK.et al. (1998Modulation of ventricular function through gene transfer in vivo Proc Natl Acad Sci USA 955251–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Grupp IL, Harrer J, Ponniah S, Grupp G, Duffy JJ.et al. (1994Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation Circ Res 75401–409. [DOI] [PubMed] [Google Scholar]
- He H, Meyer M, Martin JL, McDonough PM, Ho P, Lou X.et al. (1999Effects of mutant and antisense RNA of phospholamban on SR Ca(2+)-ATPase activity and cardiac myocyte contractility Circulation 100974–980. [DOI] [PubMed] [Google Scholar]
- Giordano FJ, He H, McDonough P, Meyer M, Sayen MR., and, Dillmann WH. Adenovirus-mediated gene transfer reconstitutes depressed sarcoplasmic reticulum Ca2+-ATPase levels and shortens prolonged cardiac myocyte Ca2+ transients. Circulation. 1997;96:400–403. doi: 10.1161/01.cir.96.2.400. [DOI] [PubMed] [Google Scholar]
- Hoshijima M, Ikeda Y, Iwanaga Y, Minamisawa S, Date MO, Gu Y.et al. (2002Chronic suppression of heart-failure progression by a pseudophosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery Nat Med 8864–871. [DOI] [PubMed] [Google Scholar]
- Chien KR, Ross J., Jr, and, Hoshijima M. Calcium and heart failure: the cycle game. Nat Med. 2003;9:508–509. doi: 10.1038/nm0503-508. [DOI] [PubMed] [Google Scholar]
- Moore M, Choo Y., and, Klug A. Design of polyzinc finger peptides with structured linkers. Proc Natl Acad Sci USA. 2001;98:1432–1436. doi: 10.1073/pnas.98.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Guschin D, Davalos A, Lee YL, Snowden AW, Jouvenot Y.et al. (2003Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity Proc Natl Acad Sci USA 10011997–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gauster M, Strauss J, Hrzenjak A., and, Kostner GM. Adenovirus-mediated apo(a)-antisense-RNA expression efficiently inhibits apo(a) synthesis in vitro and in vivo. Gene Ther. 2001;8:425–430. doi: 10.1038/sj.gt.3301434. [DOI] [PubMed] [Google Scholar]
- Huang Y, Hickey RP, Yeh JL, Liu D, Dadak A, Young LH.et al. (2004Cardiac myocyte-specific HIF-1alpha deletion alters vascularization, energy availability, calcium flux, and contractility in the normoxic heart FASEB J 181138–1140. [DOI] [PubMed] [Google Scholar]
- Reddy DS, Singh M, Ghosh S., and, Ganguly NK. Role of cardiac renin-angiotensin system in the development of pressure-overload left ventricular hypertrophy in rats with abdominal aortic constriction. Mol Cell Biochem. 1996;155:1–11. doi: 10.1007/BF00714327. [DOI] [PubMed] [Google Scholar]
- Nakayama H, Otsu K, Yamaguchi O, Nishida K, Date MO, Hongo K.et al. (2003Cardiac-specific overexpression of a high Ca2+ affinity mutant of SERCA2a attenuates in vivo pressure overload cardiac hypertrophy FASEB J 1761–63. [DOI] [PubMed] [Google Scholar]
- Suckau L, Fechner H, Chemaly E, Krohn S, Hadri L, Kockskämper J.et al. (2009Long-term cardiac-targeted RNA interference for the treatment of heart failure restores cardiac function and reduces pathological hypertrophy Circulation 1191241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaski BE, Jessup ML, Mancini DM, Cappola TP, Pauly DF, Greenberg B, Calcium Up-Regulation by Percutaneous Administration of Gene Therapy In Cardiac Disease (CUPID) Trial Investigators et al. Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (CUPID Trial), a first-in-human phase ½ clinical trial. J Card Fail. 2009;15:171–181. doi: 10.1016/j.cardfail.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID) Investigators et al. Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation. 2011;124:304–313. doi: 10.1161/CIRCULATIONAHA.111.022889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou D, Maclaughlin F, Thiesse M, Panchal VR, Bekkers BC, Wilson EA.et al. (2003Widespread regional myocardial transfection by plasmid encoding Del-1 following retrograde coronary venous delivery Catheter Cardiovasc Interv 58207–211. [DOI] [PubMed] [Google Scholar]
- Kaye DM, Preovolos A, Marshall T, Byrne M, Hoshijima M, Hajjar R.et al. (2007Percutaneous cardiac recirculation-mediated gene transfer of an inhibitory phospholamban peptide reverses advanced heart failure in large animals J Am Coll Cardiol 50253–260. [DOI] [PubMed] [Google Scholar]
- Giordano FJ. Retrograde coronary perfusion: a superior route to deliver therapeutics to the heart?*. J Am Coll Cardiol. 2003;42:1129–1131. doi: 10.1016/s0735-1097(03)00903-3. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J.et al. (2005Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart Nat Biotechnol 23321–328. [DOI] [PubMed] [Google Scholar]
- Guan X, Stege J, Kim M, Dahmani Z, Fan N, Heifetz P.et al. (2002Heritable endogenous gene regulation in plants with designed polydactyl zinc finger transcription factors Proc Natl Acad Sci USA 9913296–13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP., and, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M.et al. (2003Expression profiling reveals off-target gene regulation by RNAi Nat Biotechnol 21635–637. [DOI] [PubMed] [Google Scholar]
- Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW.et al. (2005siRNA-mediated off-target gene silencing triggered by a 7 nt complementation Nucleic Acids Res 334527–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bish LT, Sleeper MM, Reynolds C, Gazzara J, Withnall E, Singletary GE.et al. (2011Cardiac gene transfer of short hairpin RNA directed against phospholamban effectively knocks down gene expression but causes cellular toxicity in canines Hum Gene Ther 22969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Peisach E., and, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A.et al. (2003Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins Biochemistry 422137–2148. [DOI] [PubMed] [Google Scholar]
- Jamieson AC, Miller JC., and, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- Visser AE, Verschure PJ, Gommans WM, Haisma HJ., and, Rots MG. Step into the groove: engineered transcription factors as modulators of gene expression. Adv Genet. 2006;56:131–161. doi: 10.1016/S0065-2660(06)56004-3. [DOI] [PubMed] [Google Scholar]
- Moore M, Klug A., and, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci USA. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T, Elliger S, Elliger C, Podsakoff G, Villarreal L, Kurtzman GJ.et al. (1998Adeno-associated virus vectors can be efficiently produced without helper virus Gene Ther 5938–945. [DOI] [PubMed] [Google Scholar]
- Abbott JD, Huang Y, Liu D, Hickey R, Krause DS., and, Giordano FJ. Stromal cell-derived factor-1alpha plays a critical role in stem cell recruitment to the heart after myocardial infarction but is not sufficient to induce homing in the absence of injury. Circulation. 2004;110:3300–3305. doi: 10.1161/01.CIR.0000147780.30124.CF. [DOI] [PubMed] [Google Scholar]
- Lei L, Mason S, Liu D, Huang Y, Marks C, Hickey R.et al. (2008Hypoxia-inducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel-Lindau protein Mol Cell Biol 283790–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Catecholamine-O-methyltransferase expression is not regulated by the PLN repressor.
Efficiency of coinfection following intramuscular injection of two separate adenovirus vectors.
AAV-mediated delivery following direct intramyocardial injection.