For the last several decades the field of cancer genetics has really been two fields. Medical geneticists and genetic counselors use the term to describe inherited susceptibility to cancer and the identification of constitutional mutations, which convey this risk. Alternatively, many pathologists, oncologists and molecular biologists use the term to describe the multitude of genetic changes that occur in the tumor cell itself. Even among the American Board of Medical Genetics laboratory specialties, molecular diagnosticians have focused on inherited mutations, e.g. sequencing DNA from blood for tumor suppressor gene mutations whereas cytogeneticists and molecular pathologists characterize translocations, copy number changes and specific oncogenic missense mutations in tumor specimens.
This dichotomy is breaking down. Like most things in cancer genetics one can always start with retinoblastoma. The determination as to whether a patient with unilateral retinoblastoma has the hereditary form is founded on analysis of the tumor specimen to identify both RB1 inactivating events.1 This information is then used to inform analysis of the blood, e.g. if the RB1 promoter undergoes biallelic methylation then analysis of the blood isn’t indicated. The wealth of data from the Tumor Genome Anatomy project (TCGA) analysis of glioblastoma reveals that somatic mutations in susceptibility genes like RB1 are more frequent than first realized.2 As next generation sequencing becomes increasingly available practitioners of all types will need to consider both the constitutional and cancer genome when making clinical decisions. Described here are some examples of how medical geneticists are beginning to incorporate cancer genome data into clinical practice.
Analysis of relatively rare gastrointestinal tumors demonstrates the interaction between genes in the same signaling pathway. Molecular pathologists have demonstrated that a substantial portion of both hepatoblastoma and desmoid tumors contain mutations that impact WNT signaling.3 In the majority of cases these mutations are specific exon 3 missense mutations in the CTNNB1 gene encoding β-CATENIN.4 In contrast, work on hereditary tumors has demonstrated that a subset of patients with these tumors (perhaps 10-15%) harbor germline mutations in the APC gene even if other clinical features of FAP are not yet evident.5 Thus, diagnostic sequencing of the blood for mutations in APC has been recommended in all children with these tumors.5 APC and β-CATENIN proteins function in the same pathway where APC regulates the availability of β-CATENIN. More recently an inverse relationship between somatic CTNNB1 mutation and constitutional APC mutation in desmoid and hepatoblastoma tumors has been established.4;6 If the tumor contains a somatic mutation in CTNNB1 then constitutional APC mutations aren’t found. So now when I’m a referred a child with one of these tumors my first question is what did the molecular pathology of the tumor show? If one of the recurrent CTNNB1 missense somatic mutations is found then the much more expensive comprehensive analysis of APC from the blood is generally not indicated. There are many other examples of a given tumor type requiring only one event for pathway disruption. Juvenile myelomonocytic leukemia can result from inheriting a mutation in the NF1 or CBL gene or somatic mutation of the RAS signaling pathway including mutations in the PTPN11, NRAS, KRAS or ASXL1 genes.7 Large-scale genome analysis of glioblastomas demonstrate that for a given signaling pathway brain tumors contain a variety of different mutational hits but any given tumor contains on average one event that disrupts the pathway.2 Thus, we need to think about analysis of key signaling pathways involved in tumor development and distinguish when the initial lesion is an inherited mutation or results from a somatic event.
Colon cancer is one of the best examples of where population-based molecular analysis of all tumors has been initiated by geneticists, oncologists, gastroeneterologists and pathologists based on knowledge of Lynch syndrome (hereditary non-polyposis colon cancer) and the microsatellite instability (MIN) that results from defects in mismatch repair (MMR) genes. The presence of MIN or the lack of expression of one of the MMR genes detected by immunohistochemistry (IHC) is used to decide upon further molecular analyses and whether germline testing is required.8 But colon cancer also demonstrates inverse relationships between germline and somatic mutations that can’t be easily understood by simple pathway analysis. About 40% of MIN tumors harbor specific missense mutations in the BRAF oncogene.9;10 For reasons that are not clearly understood, tumors with somatic BRAF mutations almost never derive from patients with Lynch syndrome. BRAF mutations are seen in tumors with somatic methylation of MLH1 and the BRAF mutation may be an early event that is associated with abnormal methylation of multiple genes in the tumor.11 Therefore, current recommendations for molecular diagnosis of colon cancer include MIN or IHC analysis combined with BRAF mutation status to decide whether susceptibility testing is indicated. Thus the results of the tumor analysis impact prognosis and treatment decisions (e.g. 5-fluoruracil is less effective in MIN patients) while the results of germline testing impact cancer surveillance for the patient and identification of at-risk family members.11;12
Clearly, we are entering the era of comprehensive molecular analysis. This includes examining tumors for sequence changes, abnormal methylation, rearrangement (translocations and fusions) and copy number changes. The ALK proto-oncogene is an example of a single gene that undergoes multiple different mechanisms of activation in both the germline and somatically in tumors that range from neuroblastoma to lung cancer.13 Increasingly sophisticated analysis of the tumor genome will improve prediction of inherited susceptibility and will aid in distinguishing which rare constitutional variants are clinically relevant. To speed progress there needs to be better communication among the range of practitioners and researchers working on this problem such that the term “cancer genetics” has one definition.
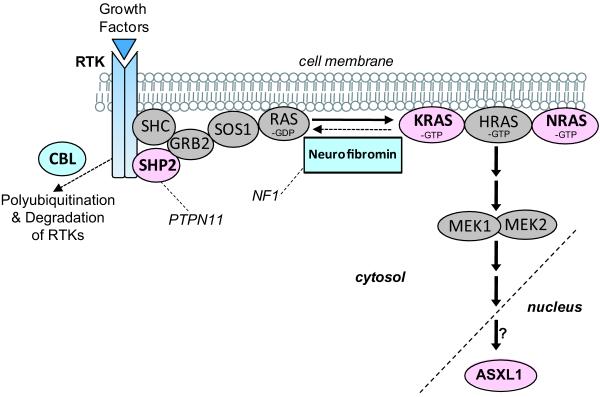
Fig. 1.
Schematic diagram of the Ras signaling pathway which is disrupted in juvenile myelomonocytic leukemia (JMML). Inherited susceptibility to JMML genes are depicted in light blue. Genes with somatic mutation in JMML are depicted in pink. The impact of Ras signaling on ASXL1 function is not clear. RTK, receptor tyrosine kinase. Modified with permission from Curr Opin Genet Dev.14
Acknowledgments
SEP was supported by grant 5R01CA138836.
Footnotes
Conflict of Interest:
The author has no commercial association that might pose or create a conflict of interest with the information presented in this manuscript. Salary support for the author was derived from a grant from the National Cancer Institute 5R01CA138836 to SEP.
REFERENCES
- 1.Rushlow D, Piovesan B, Zhang K, et al. Detection of mosaic RB1 mutations in families with retinoblastoma. Hum Mutat. 2009;30:842–851. doi: 10.1002/humu.20940. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455:1061–1068. doi: 10.1038/nature07385. 008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armengol C, Cairo S, Fabre M, Buendia MA. Wnt signaling and hepatocarcinogenesis: The hepatoblastoma model. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Terrada D, Gunaratne PH, Adesina AM, et al. Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum Pathol. 2009;40:783–794. doi: 10.1016/j.humpath.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Aretz S, Koch A, Uhlhaas S, et al. Should children at risk for familial adenomatous polyposis be screened for hepatoblastoma and children with apparently sporadic hepatoblastoma be screened for APC germline mutations? Pediatr Blood Cancer. 2006;47:811–818. doi: 10.1002/pbc.20698. [DOI] [PubMed] [Google Scholar]
- 6.Curia MC, Zuckermann M, De LL, et al. Sporadic childhood hepatoblastomas show activation of beta-catenin, mismatch repair defects and p53 mutations. Mod Pathol. 2008;21:7–14. doi: 10.1038/modpathol.3800977. [DOI] [PubMed] [Google Scholar]
- 7.Sugimoto Y, Muramatsu H, Makishima H, et al. Spectrum of molecular defects in juvenile myelomonocytic leukaemia includes ASXL1 mutations. Br J Haematol. 150:83–87. doi: 10.1111/j.1365-2141.2010.08196.x. 010. [DOI] [PubMed] [Google Scholar]
- 8.Lynch HT, Lynch JF, Lynch PM, Attard T. Hereditary colorectal cancer syndromes: molecular genetics, genetic counseling, diagnosis and management. Fam Cancer. 2008;7:27–39. doi: 10.1007/s10689-007-9165-5. [DOI] [PubMed] [Google Scholar]
- 9.Domingo E, Laiho P, Ollikainen M, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loughrey MB, Waring PM, Tan A, et al. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Fam Cancer. 2007;6:301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 11.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng M, Ott GR. Anaplastic lymphoma kinase as a therapeutic target in anaplastic large cell lymphoma, non-small cell lung cancer and neuroblastoma. Anticancer Agents Med Chem. 2010;10:236–249. doi: 10.2174/1871520611009030236. [DOI] [PubMed] [Google Scholar]
- 14.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]