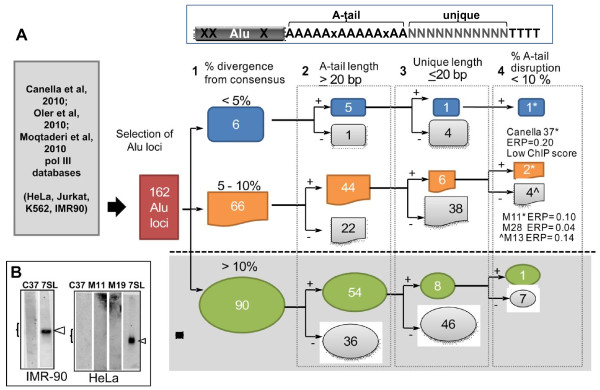
Figure 2.
Classification strategy of potentially active Alu loci. A schematic of an Alu element is on the top right, where the body is shown in gray. X represent sequences changes differing from the consensus or non-A residues in the A-tail. Ns represent the unique flanking sequence located downstream of the A-tail and the first RNA polymerase III terminator defined as four or more thymidine residues (TTTT). A. Flow chart used for the identification of potentially retrotransposition competent Alu elements. Datasets from previously published data sets were interrogated for Alu loci identified as bound by RNA polymerase III factors. A total of 162 Alu-candidate loci were retrieved and classified based on four parameters: 1- % divergence from the consensus sequence, which was subdivided into three categories based on the % sequence divergence from their Alu subfamily consensus (less than 5% in blue, 5 to 10% in orange and over 10% in green also classified as retrotranspositionally inactive). 2- A-tail length, where Alu elements with A-tail lengths of equal to or over 20 bp were included as potentially active (+). Alu elements not meeting the criteria were binned separately and not further evaluated (shown as gray symbols; -). 3- Length of the unique sequence, scoring positive Alu elements with 20 bp or less. 4- % disruption of the A-tail sequence was scored selecting for those with 10% or less disruption. The two Alu candidates evaluated for expression (show in B) are indicated by the asterisks in the flow diagram. ERP = estimated retrotranspositional potential. B. Expression of candidate Alu loci with retrotransposition potential. Total RNA extracts from HeLa or IMR90 cells were hybridized with radioactively endlabeled oligomers complementary to the unique region of the Alu loci (Additional file 1: Table S11) following a previously published protocol [24]. Results from Alu Yb8 Canella 37 (C37), Alu Sx Moq 11 h (M11), and Alu Sx Moq 19 k (M19) are shown. An oligo complementary to a region of the 7SL RNA that does not share sequence similarity to Alu RNA: 5′-CCGATCGGCATAGCGCACTA-3′ was used as a positive control (white arrowhead). Bracket approximates the expected location of the Alu transcript.