Table 1. Refinement and structure statistics of PDB entry 3vqh .
Values in parentheses are for the last shell.
| Data collection and scaling (XDS) | |
| X-ray source | ID29, ESRF |
| Resolution limits () | 35.01.95 (2.201.95) |
| Unit-cell parameters () | a = 72.73, b = 75.18, c = 80.33 |
| Space group | P212121 |
| Wavelength () | 0.91969 |
| Total No. of reflections | 283804 (84880) |
| No. of unique reflections | 61702 (18509) |
| Multiplicity | 4.6 (4.6) |
| I/(I) | 19.08 (5.68) |
| R mrgd-F † (%) | 9.0 (28.8) |
| Completeness (%) | 98.0 (99.3) |
| Wilson B (2) | 21.57 |
| Refinement (REFMAC5) | |
| R work ‡ (%) | 19.0 |
| R free ‡ (%) | 23.4 |
| Average B factor (2) | 21.88 |
| No. of protein atoms | 3015 |
| No. of ligand atoms | 62 |
| No. of water molecules | 178 |
| R.m.s.d. bond lengths () | 0.010 |
| R.m.s.d. bond angles () | 1.38 |
| Ramachandran plot (PROCHECK) | |
| Most favoured (%) | 91.1 |
| Additionally allowed (%) | 8.9 |
| Generously allowed (%) | 0 |
| Disallowed (%) | 0 |
R
mrgd-F = 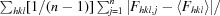
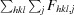 .
.
R
work/R
free = 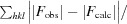
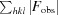 .
.
