Trigonal crystals of FadR from B. halodurans have been obtained. X-ray data were collected to 2.05 Å resolution using synchrotron radiation.
Keywords: FadR, transcriptional regulators, Bacillus halodurans
Abstract
FadR is an acyl-CoA-dependent transcription factor which regulates genes encoding proteins involved in fatty-acid degradation and synthesis in order to maintain lipid homeostasis. FadR from the alkaliphilic bacterium Bacillus halodurans was cloned and overexpressed in Escherichia coli. The FadR (Bh3102) protein from B. halodurans is composed of 195 amino-acid residues with a molecular mass of 22 378 Da. Crystals were obtained by the sitting-drop vapour-diffusion method and diffracted to 2.05 Å resolution. FadR was crystallized at 296 K using polyethylene glycol 3350 as a precipitant. The crystal belonged to the apparent trigonal space group P3221, with unit-cell parameters a = b = 56.34, c = 199.73 Å. The Matthews coefficient and solvent content were estimated to be 2.0 Å3 Da−1 and 39.8%, respectively, assuming that the asymmetric unit contained two molecules of FadR, which was subsequently confirmed by molecular-replacement calculations.
1. Introduction
Fatty acids are essential biomolecular components for all living organisms; they are vital components of the cell membrane and can be used as important energy sources for metabolism. Therefore, the degradation and biosynthesis of fatty acids are strictly regulated in terms of lipid homeostasis based on the availability of extracellular fatty acids (Xu et al., 2001 ▶). FadR is a main transcription factor for the regulation of genes involved in fatty-acid degradation and synthesis pathways. The FadR regulation pathway has been extensively studied in two model prokaryotes: Escherichia coli and Bacillus subtilis (Fujita et al., 2007 ▶).
E. coli FadR, which is a GntR-family transcriptional regulator, negatively regulates genes associated with the fatty-acid transport system (fadL and fadD), enzymes of the β-oxidation cycle (FadA, FadB, FadE, FadF, FadG and FadH), glyoxylate shunt enzymes (AceA, AceB and AceK) and the universal stress protein UspA (DiRusso et al., 1999 ▶; Farewell et al., 1996 ▶; Gui et al., 1996 ▶). FadR is released from the operator by interaction with long-chain (C14–C18) fatty-acyl-CoAs, resulting in derepression of the fad genes (Henry & Cronan, 1991 ▶, 1992 ▶; DiRusso et al., 1992 ▶). E. coli FadR also positively regulates fab genes involved in unsaturated fatty-acid biosynthesis. FadR activates the fabA and fabB genes repressed by FabR, which functions as a repressor of fabB and fabA expression, resulting in induction of fatty-acid biosynthesis (Henry & Cronan, 1991 ▶, 1992 ▶; Campbell & Cronan, 2001 ▶). Crystal structures of E. coli FadR have been determined in three different forms: an apo form, an acyl-CoA-bound form and a DNA complex. They revealed that it functions as a homodimer and that each subunit is composed of an N-terminal DNA-binding domain and a C-terminal acyl-CoA-binding domain (van Aalten et al., 2000 ▶, 2001 ▶; Xu et al., 2001 ▶).
B. subtilis FadR, which belongs to the TetR-family transcriptional regulators and is a functional homologue of E. coli FadR, represses a considerable number of genes involved in fatty-acid degradation, such as lcfA, fadB, eftA, eftB, lcfB, fadB, fadA, fadE, fadH and acdA. In a similar manner to E. coli FadR, B. subtilis FadR is deactivated by long-chain fatty-acyl-CoAs (Matsuoka et al., 2007 ▶). However, B. subtilis FadR has not been reported to activate fatty-acid biosynthesis. In B. subtilis, FapR negatively regulates the expression of the fap regulon (fabHA-fabF, fapR-plsX-fabD-fabG, fabI, fabHB, yhfX and plsC) involved in biosynthesis of fatty acids and phospholipids (Campbell & Cronan, 2001 ▶). The crystal structure of B. subtilis FadR revealed that it was a homodimer and that one lauroyl-CoA molecule was incorporated at the C-terminal acyl-CoA-binding domain in one of the subunits (Badger et al., 2005 ▶). In contrast, the crystal structure of Thermus thermophilus FadR showed that each subunit contained a lauroyl-CoA molecule (Agari et al., 2011 ▶). The acyl-CoA-binding mode of B. subtilis FadR is similar to that of T. thermophilus FadR, which also belongs to the TetR family, but is quite different from that of E. coli FadR.
The FadR homologue (Bh3102) from B. halodurans encodes a protein of 195 amino-acid residues with 65% sequence identity to B. subtilis FadR and 21% sequence identity to T. thermophilus FadR. Although the crystal structure of B. subtilis FadR was determined in a structural genomics project (Badger et al., 2005 ▶), a detailed study of its structure has not yet been reported. It would be attractive to determine the structure of the FadR orthologue from B. halodurans. As a first step, we report the cloning, purification, crystallization and preliminary X-ray diffraction data of B. halodurans FadR.
2. Materials and methods
2.1. Expression and purification
The gene encoding FadR (Bh3102) was amplified by polymerase chain reaction using genomic DNA of B. halodurans as a template. It was inserted into the NdeI/XhoI-digested expression vector pET-28b(+) (Novagen) to produce protein with a hexahistidine tag at the N-terminus. The recombinant B. halodurans FadR protein was expressed in E. coli BL21 (DE3) Star pLysS cells (Invitrogen). The cells were grown at 310 K to an OD600 of ∼0.5 in Luria–Bertani medium supplemented with 30 µg ml−1 kanamycin and 30 µg ml−1 chloramphenicol. Protein expression was induced using 1.0 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cell growth was continued at 303 K for 4 h after IPTG induction and the cells were harvested by centrifugation at 4200g for 10 min at 277 K.
The cell pellet was resuspended in lysis buffer [20 mM Tris–HCl pH 8.0, 0.5 M NaCl, 10%(v/v) glycerol, 1 mM phenylmethylsulfonyl fluoride] and homogenized using an ultrasonic processor. The insoluble fraction including cellular debris was removed by centrifugation at 31 000g for 60 min at 277 K and the recombinant protein in the supernatant fraction was purified using three chromatographic steps. The first step was metal-chelate chromatography on Ni–NTA resin (GE Healthcare). The His-tagged FadR protein was eluted with buffer A [20 mM Tris–HCl pH 8.0, 0.5 M NaCl, 10%(v/v) glycerol] containing 300 mM imidazole followed by enzymatic removal of the His tag by overnight incubation with PreScission protease. Uncleaved His-tagged protein and PreScission protease were removed from the target FadR by application onto an Ni–NTA affinity column. The next step was gel filtration on a Superdex 75 column (GE Healthcare) employing an elution buffer consisting of 0.2 M NaCl, 20 mM Tris–HCl pH 8.0, 1 mM DTT, 5 mM MgCl2, 5%(v/v) glycerol. The purified protein was concentrated to 70 mg ml−1 using Centricon YM-10 (Millipore) and aliquots of the protein were stored at 193 K.
2.2. Crystallization
Crystallization screening was initially set up using a Mosquito crystallization robot (TTP LabTech) using concentrated FadR and various commercial screening solution kits from Hampton Research, Qiagen and Emerald BioSystems. Crystals were obtained by the sitting-drop vapour-diffusion method using 96-well CrystalQuick plates (Greiner Bio-One); each crystallization drop was prepared by mixing 0.2 µl reservoir solution and 0.2 µl protein solution. Initial crystals were obtained in several conditions containing polyethylene glycol (PEG) 3350 and magnesium chloride, which were further optimized to 25%(v/v) PEG 3350, 0.1 M Tris–HCl pH 8.5, 0.3 M magnesium chloride.
2.3. X-ray data collection
Crystals were transferred into a cryoprotectant solution consisting of 20%(v/v) glycerol in the reservoir solution and immediately flash-cooled in liquid nitrogen. X-ray diffraction data were collected at 100 K on an ADSC Quantum 210 CCD image-plate detector using synchrotron radiation on beamline BL-5A of the Photon Factory, Japan. The data were collected using 1° oscillation per image with a crystal-to-detector distance of 200 mm. The crystal was exposed to X-rays for 1 s per image and a total of 200 frames were recorded. Data were processed and scaled using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
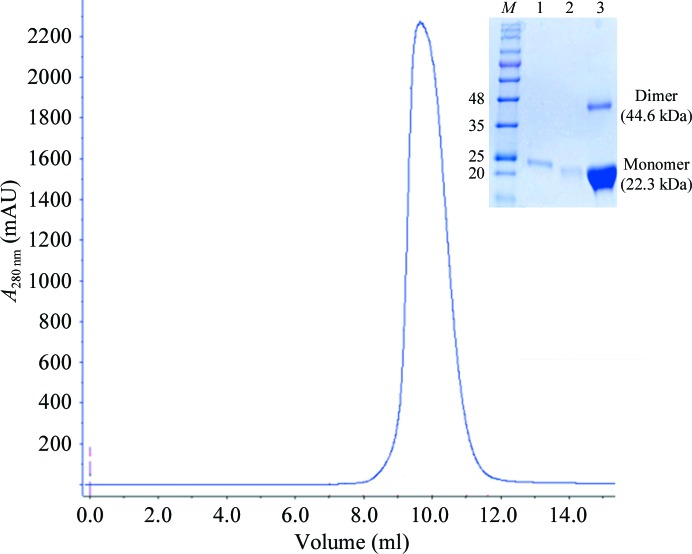
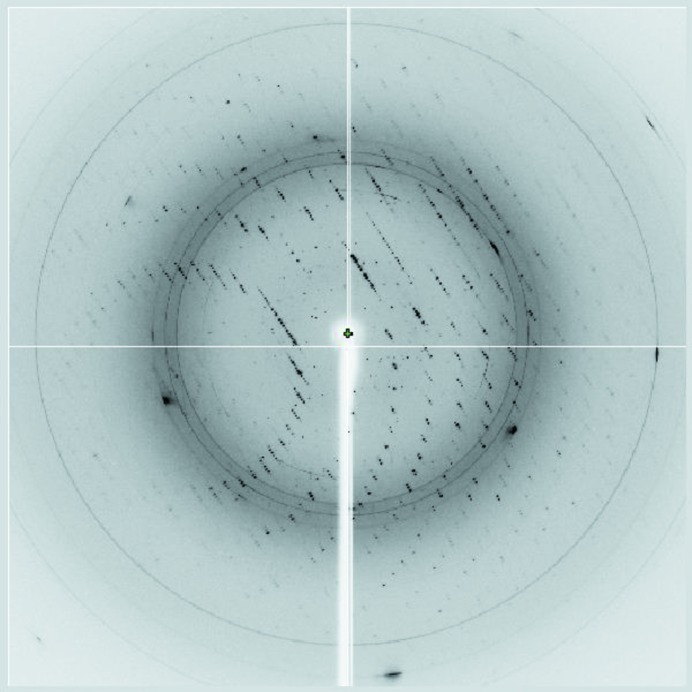
Recombinant B. halodurans FadR protein was expressed in E. coli and purified to give a final yield of ∼50 mg per litre of culture (Fig. 1 ▶). Reproducible crystals were obtained by sitting-drop vapour diffusion at 296 K from optimized reservoir solution consisting of 25%(v/v) PEG 3350, 0.1 M Tris–HCl pH 8.5, 0.3 M magnesium chloride and grew to approximate dimensions of 0.4 × 0.4 × 0.15 mm within a week (Fig. 2 ▶). The best crystal was transferred into cryoprotectant solution consisting of the reservoir solution with 20%(v/v) glycerol. A crystal was flash-cooled in liquid nitrogen and mounted at the experimental station on beamline BL-5A of the Photon Factory. A total of 24 120 unique reflections were measured with a multiplicity of 6.7 (Fig. 3 ▶). The merged data set was 98.6% complete to 2.05 Å resolution and gave an R merge (on intensities) of 5.9%. The crystals belonged to a primitive trigonal space group, with unit-cell parameters a = b = 56.34, c = 199.7 Å. Systematic absences of reflections indicated that the space group could be P3221 or P3121. The asymmetric unit contained two molecules of FadR, giving a crystal volume per mass (V M) of 2.0 Å3 Da−1 and a solvent content of 39.8% (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶). Table 1 ▶ summarizes the the statistics of data collection.
Figure 1.
Purification of B. halodurans FadR. Isocratic elution chromatogram of the recombinant protein separated by gel-filtration chromatography on a Superdex 75 column. Inset, 12% SDS–PAGE stained with Coomassie Blue; lane M, molecular-weight markers (labelled in kDa); lane 1, His-tagged FadR eluted from the Ni column; lane 2, eluate from the Ni column after PreScission protease digestion; lane 3, eluate from the Superdex 75 column.
Figure 2.

Trigonal crystals of B. halodurans FadR. Their approximate dimensions are 0.4 × 0.4 × 0.15 mm.
Figure 3.
X-ray diffraction image from a crystal of B. halodurans FadR. The diffraction image was obtained on beamline BL-5A at the Photon Factory. The crystal-to-detector distance was 200 mm and the wavelength was 1.000 Å.
Table 1. Data-collection statistics for the B. halodurans FadR crystal.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.000 |
| Temperature (K) | 100 |
| Oscillation range (°) | 1 |
| Resolution range (Å) | 20.0–2.05 (2.09–2.05) |
| No. of observations | 157996 |
| Unique reflections | 23715 |
| Space group | P3221 |
| Unit-cell parameters (Å) | a = b = 56.34, c = 199.7 |
| Data completeness (%) | 98.6 (100.0) |
| Multiplicity | 6.7 (6.9) |
| Average I/σ(I) | 39.6 (7.8) |
| R merge † (%) | 5.9 (32.6) |
R
merge = 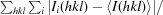
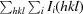 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl, 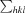 is the sum over all reflections and
is the sum over all reflections and 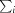 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
Molecular replacement was attempted with the program Phaser (McCoy et al., 2007 ▶) using the B. subtilis FadR structure (PDB entry 1vi0; 61% sequence identity; Badger et al., 2005 ▶). Two FadR monomers were successfully located in the asymmetric unit and the correct space group was determined to be P3221 from the two possible space groups. Model building and structure refinement are under way.
Acknowledgments
We thank the staff members of BL-5A at the Photon Factory for their help with data collection. We are grateful to Drs Hye-Jin Yoon and Hyun Suk Kim for data collection. This work was supported by a National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011-0014251).
References
- Aalten, D. M. F. van, DiRusso, C. C. & Knudsen, J. (2001). EMBO J. 20, 2041–2050. [DOI] [PMC free article] [PubMed]
- Aalten, D. M. F. van, DiRusso, C. C., Knudsen, J. & Wierenga, R. K. (2000). EMBO J. 19, 5167–5177. [DOI] [PMC free article] [PubMed]
- Agari, Y., Agari, K., Sakamoto, K., Kuramitsu, S. & Shinkai, A. (2011). Microbiology, 157, 1589–1601. [DOI] [PubMed]
- Badger, J. et al. (2005). Proteins, 60, 787–796. [DOI] [PubMed]
- Campbell, J. W. & Cronan, J. E. (2001). J. Bacteriol. 183, 5982–5990. [DOI] [PMC free article] [PubMed]
- DiRusso, C. C., Black, P. N. & Weimar, J. D. (1999). Prog. Lipid Res. 38, 129–197. [DOI] [PubMed]
- DiRusso, C. C., Heimert, T. L. & Metzger, A. K. (1992). J. Biol. Chem. 267, 8685–8691. [PubMed]
- Farewell, A., Diez, A. A., DiRusso, C. C. & Nyström, T. (1996). J. Bacteriol. 178, 6443–6450. [DOI] [PMC free article] [PubMed]
- Fujita, Y., Matsuoka, H. & Hirooka, K. (2007). Mol. Microbiol. 66, 829–839. [DOI] [PubMed]
- Gui, L., Sunnarborg, A. & LaPorte, D. C. (1996). J. Bacteriol. 178, 4704–4709. [DOI] [PMC free article] [PubMed]
- Henry, M. F. & Cronan, J. E. (1991). J. Mol. Biol. 222, 843–849. [DOI] [PubMed]
- Henry, M. F. & Cronan, J. E. (1992). Cell, 70, 671–679. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Matsuoka, H., Hirooka, K. & Fujita, Y. (2007). J. Biol. Chem. 282, 5180–5194. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Xu, Y., Heath, R. J., Li, Z., Rock, C. O. & White, S. W. (2001). J. Biol. Chem. 276, 17373–17379. [DOI] [PubMed]