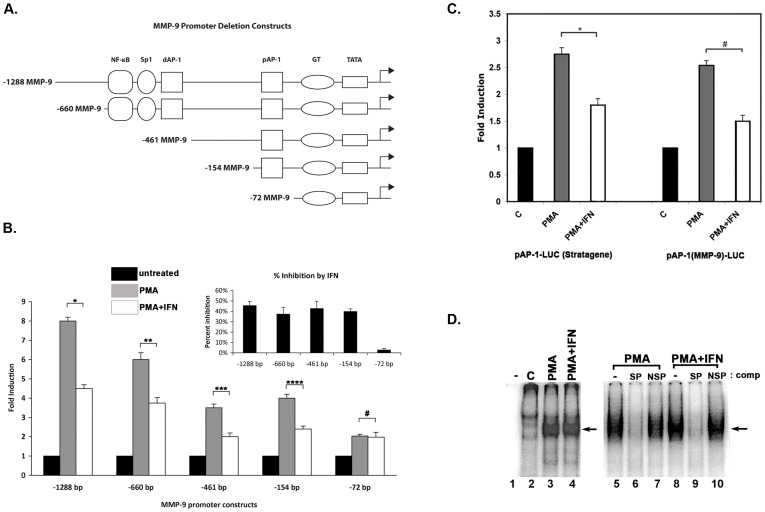
Figure 1. The -154 bp promoter construct retains the ability to respond to IFNβ.
(A) Schematic diagram of the deletion constructs. Progressive deletions of MMP-9 promoter were created by PCR amplification with appropriate primers. Various transcription factor binding sites and the TATA box are as indicated. (B) Mapping the promoter region responsible for IFNβ mediated transcriptional repression. HT1080 cells were transiently transfected in 6 well plates in triplicates with MMP-9 promoter deletion constructs as indicated using Effectene transfection reagent (Qiagen). A plasmid encoding Renilla luciferase (pRL-null, Promega) was co-transfected for the normalization of transfection efficiencies. 24 hours after transfection, the cells were treated with 30 nM PMA and 100 U/mL IFNβ for 18 hours prior to preparation of cell extracts. Cell extracts were assayed for firefly and Renilla luciferase activities. Black bars indicate untreated values, considered as 1 for each construct. Grey bars indicate PMA treated values represented as fold inductions. White bars indicate PMA and IFNβ treated values also represented as fold inductions. The values represent averages of triplicate samples from four different experiments for each construct and the error bars represent standard deviation. The P-values were calculated using statistical analysis software. As indicated above bars, *, **, ***, and **** symbols indicate P values that indicate significant difference (0.0009, 0.0007, 0.0008, and 0.0009 resp.) and the symbol # indicates a P value of 0.1216 (no significant difference) with significant values being <0.01. Inset: The percent inhibition in response to IFNβ is plotted for each deletion construct. (C) AP-1 binding site is sufficient to confer downregulation by IFNβ on a synthetic reporter construct. HT1080 cells were co-transfected with either consensus AP-1 binding site concatamer (pAP-1-Luc, Stratagene) or MMP-9-specific AP-1 binding site concatamer (AP-1 (MMP9)-Luc) linked to a basal promoter-Luc, and a plasmid encoding Renilla luciferase (pRL-null, Promega). 24 hours after transfection, cells were treated with 30 ηM PMA and 100 U/mL IFNβ for 18 hours prior to preparation of cell extracts. Cell extracts were assayed for firefly and Renilla luciferase activities. Black bars indicate untreated cell values, considered as 1 for each construct. Grey bars indicate the PMA treated values represented as fold inductions. White bars indicate the PMA and IFNβ treated values represented as fold inductions. The values are averages from three separate experiments and the error bards represent standard deviation. The P-values were calculated using statistical analysis software. As indicated above the bars, *symbol indicates a P value of 0.0008 (significant difference) and the symbol # indicates a P value of 0.0001 (significant difference), with significant values being <0.01. (D) IFNβ does not inhibit the DNA-binding activity of AP-1. EMSA was carried out using the nuclear extracts prepared from HT1080 cells that were untreated or treated with PMA, PMA plus IFNβ for 30 minutes. 1 µg of nuclear extracts were incubated with the 32P-labeled AP-1 (region −93 to −56 of MMP-9 promoter) probe. Arrows indicate AP-1 complex bound to DNA. Unlabelled specific (SP: AP-1 consensus sequence) and non-specific (NSP: TATA box consensus sequence) competitor oligonucleotides in 100-fold molar excess were utilized to confirm the identity of the bound complex.