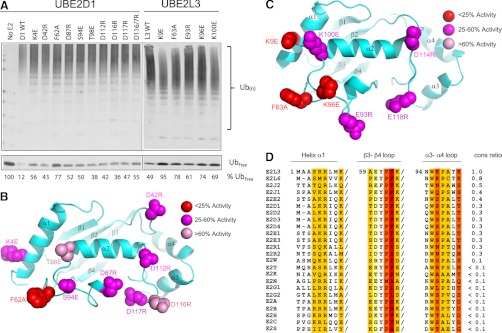
Fig. 4.
Defining residues important for E2-HECT function. A, Mutational analysis of the UBE2D1 acidic trough and UBE2L3 HECT binding domain. (top) E2 proteins bearing single point mutations (as indicated) were assayed in a standard autoubiquitylation assay. Percentage unconjugated (free) Ub is indicated below each reaction. (bottom) B, UBE2D1 ribbon diagram highlighting the locations of each of the mutated residues in the E2 structure, color coded according to magnitude of effect on Ub conjugation in vitro. C, UBE2L3 ribbon diagram highlighting locations of mutated residues. D, Alignment of the previously defined UBE2L3 HECT binding domain with other Ub-loading human E2 sequences. Residues previously found to play a role in HECT binding are highlighted. Column color corresponds to the change in binding energy observed when this residue was mutated (39). A “conservation score” for each E2 was assigned as follows; identical residues were assigned a score according to binding energy, where: red residues = +4, orange = +3, mustard = +2, light yellow = +1; conserved mutations at the same location (e.g. R to K, or S to T) were assigned half scores. Similar amino acids (e.g. aliphatic, charged or small amino acids) were assigned a score of 0. Dramatically altered amino acids at the same position were penalized with a negative score corresponding to the binding energy of each UBE2L3 amino acid. Raw scores were summed for each E2, and conservation score determined by dividing by the E2L3 score of 32.