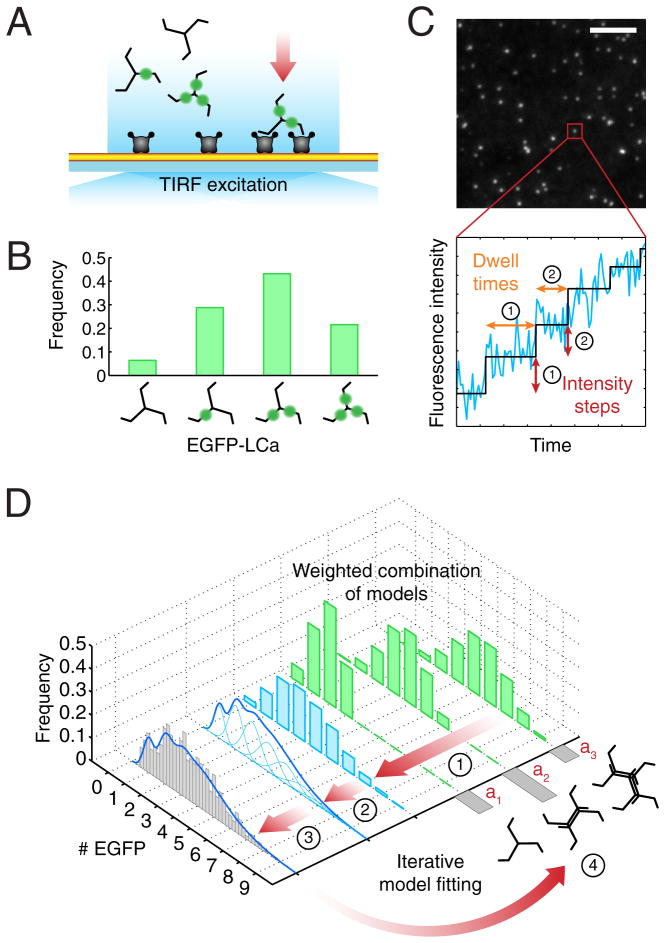
Figure 1. Schematic representation of the experimental setup and computational analysis.
(A) TIRF imaging setup used for the experiments. This illustration shows an example of the recruitment of clathrin and AP2 adaptors to the plasma membrane (yellow) of a cell attached through the modified PEG support to a glass coverslip. Each clathrin triskelion contains three light chains that, depending on the expression level, might or not be replaced by ectopically expressed EGFP-LCa (green). The imaging experiments used here were designed to detect and track with single molecule sensitivity the arrival of the first clathrin (or AP2) molecules to the site of coated pit initiation.
(B) Distribution of numbers of triskelions containing none, one, two or three EGFP-LCa molecules. Incorporation of EGFP-LCa follows a binomial distribution, shown in this example for a replacement of 60%.
(C) Frame from a TIRF time-series showing fluorescent spots corresponding to coated pits labeled with clathrin EGFP-LCa. Scale bar: 5 μm. The fluorescence intensity trace (blue) corresponds to the profile from a single clathrin coated pit imaged during its initiation phase. A step-fitting function (black) was used to determine the fluorescence intensity and dwell time (duration) associated with the first two steps (indicated by their respective numbers); these values were combined to generate the fluorescence intensity distribution shown in the gray histogram of panel (D).
(D) Data flow and computational analysis. Stage 1 shows the weighted contributions (green) for the calculated distribution of the number of EGFPs corresponding to different modes of triskelion recruitment (weights a1 to a3) constrained by the known molecular structures. For example, in the case of clathrin, EGFPs can only arrive in sets of one, two, three, etc. as part of one or more triskelions, each containing either zero, one, two or three EGFP-LCa, their relative amounts weighted by the extent of light chain replacement (panel B). Stage 2 shows the combined weighted distribution of numbers of EGFPs (blue) from stage 1 and the calculated EGFP distribution obtained by convolving the distribution of calculated EGFP molecules with Gaussians (light blue), for which the means and standard deviations were derived from the single-molecule calibration of fluorescence intensity. Calibrations are presented in Figure S1. Stage 3 corresponds to the comparison between the calculated EGFP intensity distribution (blue trace) and the experimentally determined EGFP measurements (gray). The number of clathrin triskelions recruited during a step is obtained by an iterative search process that minimizes the least-squares difference of the cumulative distribution functions between the experimentally and the calculated EGFP distribution values yielding the final weighted contribution and light chain substitution parameters (Stage 4).