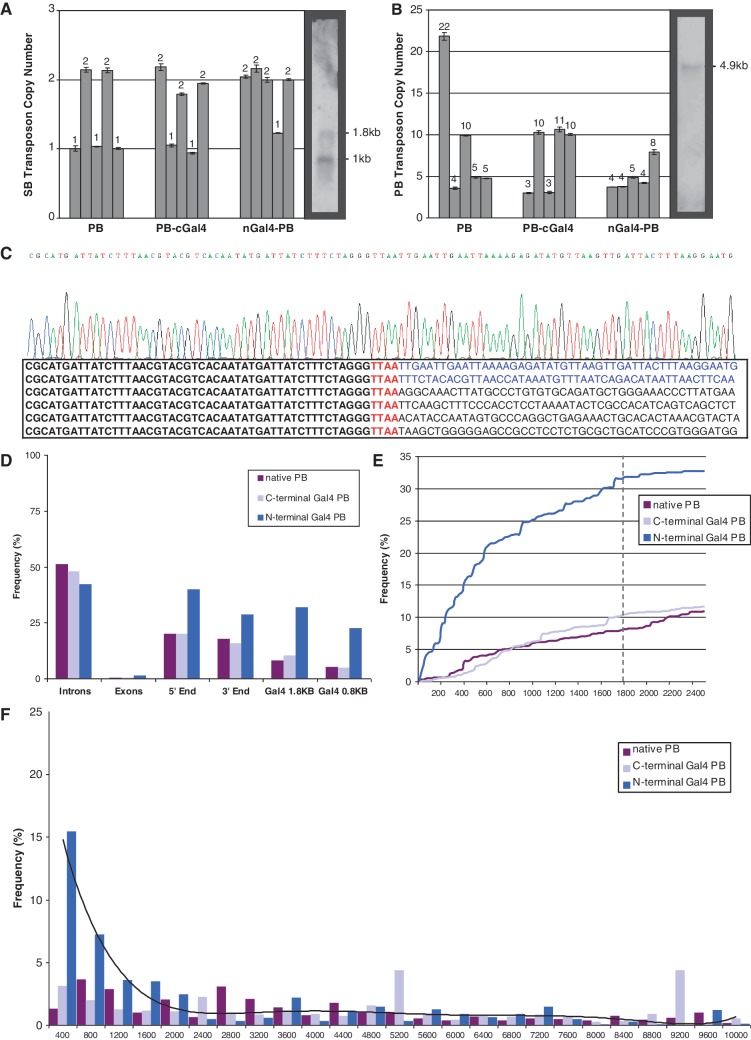
Figure 3.
(A and B) Copy number assays for number of SB and PB transposon integrations. The gDNA from 5 single clones each for N- and C-terminal Gal4 fusion and native PB experiments was analyzed by duplex Taqman real-time PCR. The numbers above the bars represent the estimated copy number for each sample. The Southern blot shown on the right of each graph was applied as a standard of known number of transposon integrations and was used to calibrate the qPCR data. (C) Sequences recovered from a representative sample showing the PB TRE on the left in bold, TTAA and flanking sequence on the right. The top 2 lines with flanking sequence in blue show nested PCR products that align to the genomic UAS–SB recipient transposon. The bottom four lines with flanking sequence in black show recovered nrLAM and 454 sequences representing off-target events with alignments to various locations in the human genome. (D) The frequency of insertion sites recovered from nrLAM PCR that land within introns and exons, within a 10 kb window surrounding transcriptional start sites (5′-end) or polyA termination sites (3′-end), and ±1.8 kb and ±0.8 kb of endogenous Gal4 recognition sites. (E) The cumulative percentage of total integrations from 0 to 2400 bp from endogenous recognition sequences. The frequency of insertions for native PB and PB–cGal4 increased linearly. nGal4–PB insertion frequency increased logarithmically until 1800 bp and then increased linearly. (F) Histogram displaying the percentage of total integrations that occurred within 400 bp intervals from 0 to 10 000 bp from endogenous Gal4 recognition sequences. The black line represents the best fit curve for nGal4–PB.