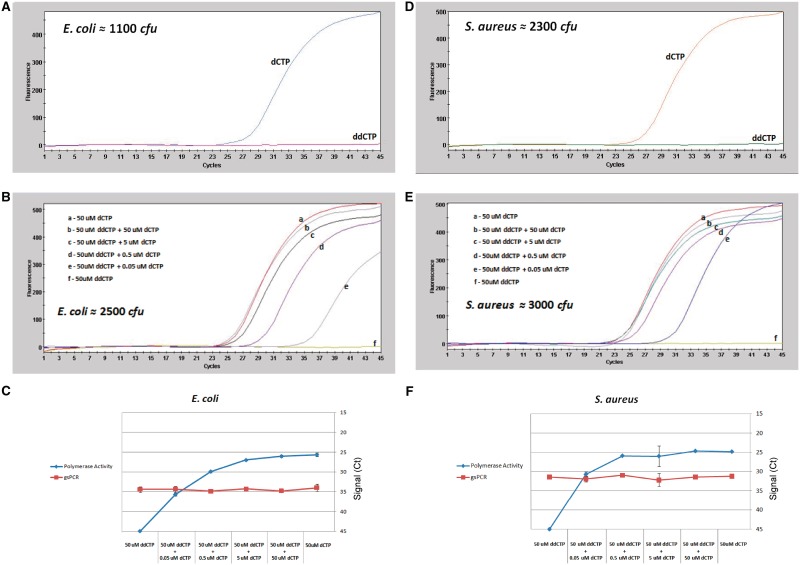
Figure 5.
Detection of bacteria by DPE-PCR is blocked by ddCTP and rescued with dCTP. (A) E. coli suspensions were added to bead lysis-coupled DNA polymerase assays composed of a 50 µM (dATP, dGTP, dTTP) mixture supplemented with either 50 µM dCTP or 50 µM ddCTP. DPE-PCR curves representing E. coli-derived DNA polymerase activity is presented. Approximate colony forming unit input as determined by plating is presented in the upper left region of the qPCR graph (B) E. coli suspensions were added to bead lysis tubes containing 50 µl reaction buffer with 50-µM (dATP, dGTP, dTTP, ddCTP). Prior to lysis, 1 µl of dCTP (2.5, 0.25, 0.025 and 0.0025 mM) was added to selected ddCTP-containing reactions. Reactions containing 50 µM (dATP, dGTP, dTTP, dCTP) alone or 50 µM (dATP, dGTP, dTTP, ddCTP) alone were run in parallel as ‘non-terminated’ and ‘terminated’ comparators. The resultant DPE-PCR curves representing E. coli-derived DNA polymerase activity is presented. Approximate colony forming unit input as determined by plating is presented in the lower left region of the qPCR graph. (C) Escherichia coli gene-specific PCR was also performed on the same lysates used for DNA polymerase detection presented in Figure 2B. Linear plots of dCTP-dependent rescue of bacterial DNA polymerase detection versus gsPCR of genomic DNA are shown. Plots were generated using the average qPCR Ct values from triplicate reactions at the indicated conditions. (D–F) ddCTP termination and dCTP rescue experiments were performed for S. aureus exactly as described above for E. coli.