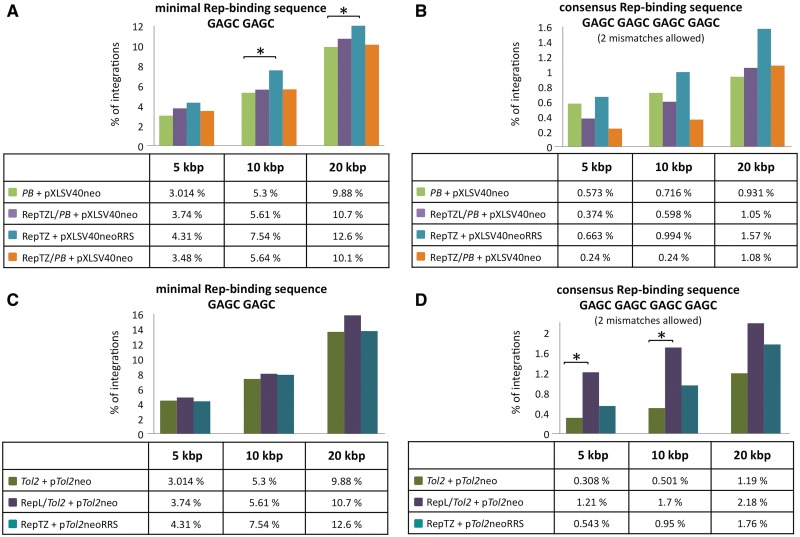
Figure 9.
Mapping of PB and Tol2 transposon integrations in proximity to chromosomal Rep-binding motifs. Transposon insertions (A and B, PB; C and D, Tol2) were recovered from HeLa cells transfected with components of the Rep-based targeting systems. Sequences neighbouring the insertion sites were analysed with respect to proximity to the next Rep-binding site. Calculations were performed with the following putative Rep-binding sites: (A and C) minimal Rep-binding site GAGC GAGC and (B and D) consensus Rep-binding site GAGC GAGC GAGC GAGC with up to two mismatches. Distances were categorized in 5-, 10- and 20-kb windows around putative Rep-binding sites and the percentage of insertions obtained in the presence and absence of the targeting fusion proteins in each category are shown. The levels of significance are marked by asterisks. P-values > 0.05 were not considered statistically significant. P-values < 0.05 (*) were considered significant. The PB data set was composed of 1397 integration sites generated with the unmodified PB transposase (PB + pXLSV40neo), 834 integration sites generated with the RepTZ/PB chimeric transposase (RepTZ/PB + pXLSV40neo) and 1207 integration sites generated with the RRS-containing PB transposon in the presence of the Rep DNA-binding protein (RepTZ + pXLSV40neoRRS). The Tol2 integration site analyses comprised 2595 integrations generated with the unmodified Tol2 transposase (Tol2 + pTol2neo), 412 integrations generated with the chimeric RepL/Tol2 transposases and 737 integrations generated in the presence of the Rep DNA-binding protein and a RRS-containing Tol2 transposon (Rep/TZ + pTol2neoRRS).