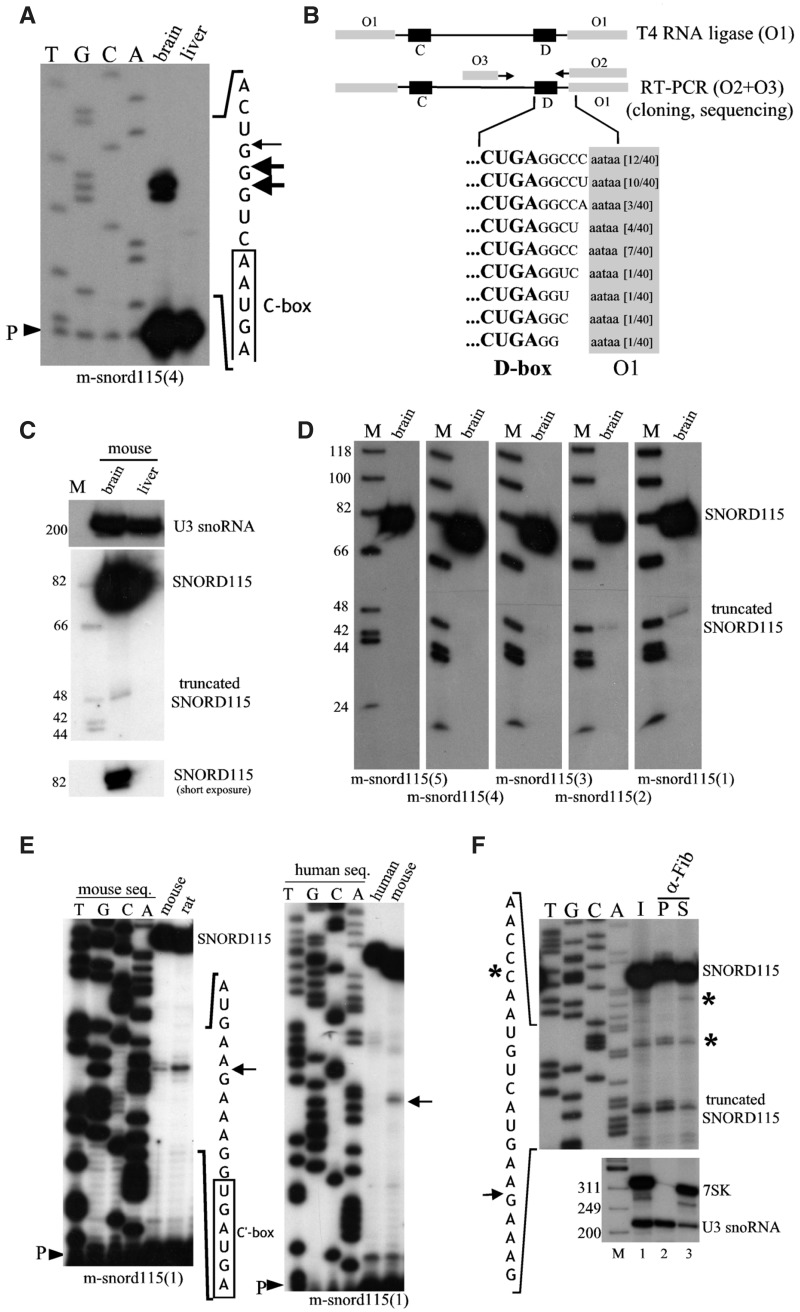
Figure 2.
The major cellular form of SNORD115 possesses a canonical 5′–3′ terminal stem structure and generates a truncated C′–D box small RNA. (A) Identifying the 5′-ends of SNORD115. Primer extension with 32P-labeled oligoprobe m-snord115(4) using 10 µg of total RNA extracted from brain or liver (negative control). P: primer. (B) Identifying the 3′-ends of SNORD115. Mouse brain total RNA was oligonucleotide-tagged at both ends (primer O1) and amplified by RT-PCR (primers O2 and O3). The numbers of sequenced clones are indicated in brackets, sequences of the 3′ arm are highlighted in grey. (C) Northern-blot hybridization to internally 32P-labeled antisense mouse SNORD115 riboprobes using 10 µg of total RNA extracted from brain or liver as indicated above the panels. Two exposures (long and short) are shown for SNORD115. U3 snoRNA was used as a gel loading control. (D) Northern-blot hybridization to 5′-32P end-labeled DNA oligonucleotides (sequences are given in Figure 1B) using 10 µg of total mouse brain RNA. (E) 5′-end identification of the 48-nt-long SNORD115-derived small RNA by primer extension with 32P-labeled snord115(1) using 10 µg of total RNA extracted from rat and mouse brains (left panel) or human and mouse brain (right panel). P: primer. (F) Immunoprecipitation by anti-fibrillarin antibodies (72B9, a generous gift of Dr. M. Pollard). Top panel: full-length SNORD115 and its 5′-truncated form were detected by primer extension as indicated in (E). The asterisks denote signals not reproducibly detected and that most likely correspond to RNA degradation that occurs upon brain extract preparation. I: Input (1/10); S: Supernatant (1/10); P: Pellet. Bottom panel: The same RNA samples were also assayed by Northern-blot analysis with U3- and 7SK-specific oligo-probes used as positive and negative controls, respectively. M: DNA marker (nt).