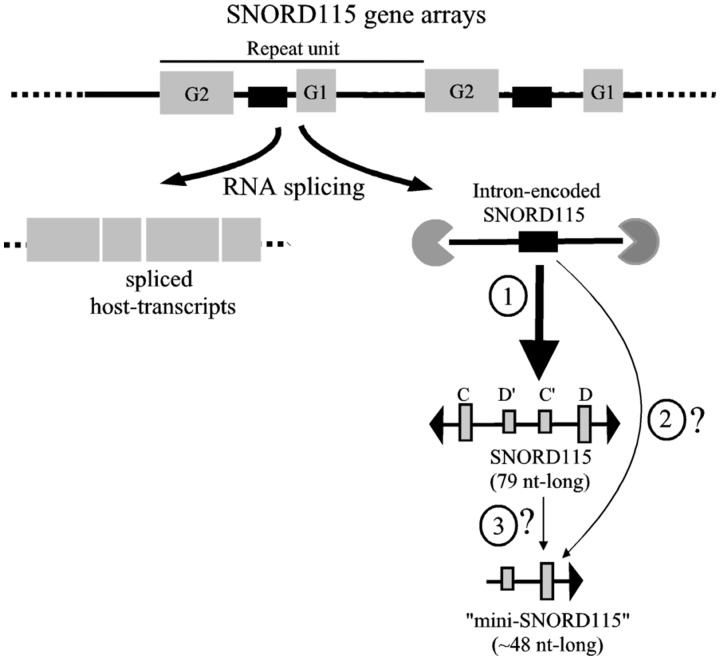
Figure 4.
Revisiting snoRNP biogenesis at the Snurf–Snrpn domain. The major cellular RNA form generated at the SNORD115 gene arrays from the Snurf–Snrpn domain corresponds to a canonical box C/D snoRNAs, most likely produced from exonucleolytic trimming of the debranched, spliced-out intron, as assumed for other intron-encoded snoRNAs (47) (pathway 1). The same holds true for SNORD116 gene arrays (data not shown). SNORD115 also generates low-abundance 5′-truncated C′–D snoRNPs. This may occur during the nucleoplasmic snoRNP assembly events before, or concomitantly with, RNA splicing of the host intron, via a minor (or aberrant) alternative snoRNA-processing pathway (pathway 2). Alternatively, C′/D snoRNPs, whose functional relevance—if any—remains to be demonstrated, may also derive from functional, fully processed snoRNPs, in the course of snoRNP remodeling and/or snoRNP decay (pathway 3).