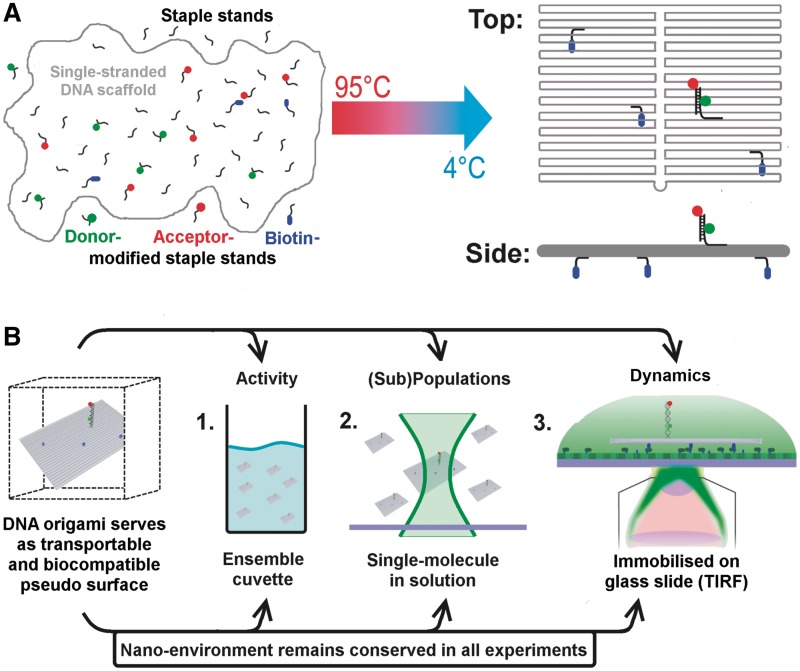
Figure 1.
Schematic drawing of the experimental strategy that allows a direct comparison of ensemble and single-molecule experiments as the DNA origami provides an identical nano-environment in all experiments. (A) DNA origami structures are based on a ‘scaffold’ DNA strand (the single-stranded DNA genome of bacteriophage M13), which can be folded into 2D and 3D assemblies at the nanometre scale with the help of hundreds of short oligonucleotides called ‘staple strands’ (24). DNA origami represent a self-assembled system; the formation of the DNA nanostructure is achieved by a simple heat denaturation step followed by slowly cooling down the scaffold DNA/staple strands mixture. Modifications (e.g. biotin, fluorophores) can be introduced into the DNA origami by replacing individual staple strands with a modified version of the oligonucleotide. (B) The decorated DNA origami represents a transportable pseudo-surface for a biomolecular reaction that can be used in a range of fluorescence-based methods. Importantly, the DNA origami ensures an identical nano-environment for the biomolecular assay (shown here is the fluorescently labelled double-stranded TATA–box containing oligonucleotide) leading to comparable reaction conditions. If biotins are positioned at one ‘face’ of a rectangular DNA origami (blue spheres) an oriented immobilization of the rectangle on a streptavidin-covered glass surface becomes possible. Consequently, the opposite side of the DNA rectangle that faces away from the surface is available for the DNA sequences that mediate the specific interaction for biomolecule immobilization, e.g. via protein–DNA interactions.