Figure 1.
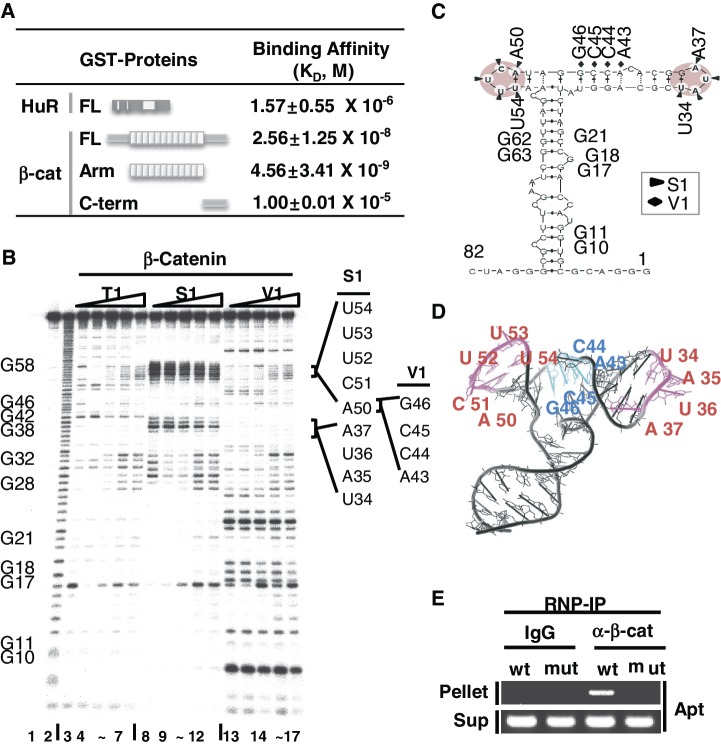
Structural probe for β-catenin-binding site mapping of the RNA aptamer. (A) Determination of in vitro equilibrium dissociation constants, KD, for the RNA aptamer by surface plasmon resonance (SPR) analysis. Schematic diagrams of HuR and β-catenin, FL, Arm and C-term of β-catenin, are shown. KD of FL, Arm and C-term of β-catenin to RNA aptamer were compared to that of HuR binding to the RNA aptamer. RNA was immobilized on a SPR chip and analyte proteins were added. Three independent SPR experiments were performed and the average KD values are shown with the standard deviation. (B) RNase mapping and footprinting of the β-catenin-binding RNA aptamer. The RNA aptamer was end-labeled and incubated with various concentrations of the β-catenin protein, followed by digestion with RNase T1, RNase S1 or RNase V1. Lane 1, no treatment; lane 2, alkaline hydrolysis; lanes 3, 8, 13, RNase treated, no β-catenin protein added; lanes 4–7, 9–12, 14–17, RNase treated with RNA–protein at ratios of 1:1, 1:2, 1:5, 1:10. G nucleotide positions are indicated on the left side of the gel. RNase-protected nucleotides (UAUA and ACUUU) are marked by solid lines on the right side of the gel. (C) MC-fold predicted secondary structure of the RNA aptamer. RNase protected sequences are indicated by triangles (S1) or diamonds (V1) and putative β-catenin-binding stem–loops are shaded. (D) MC-fold predicted tertiary structure of the aptamer was drawn using Pymol. S1 protected single-stranded ARE sequences (magenta, UAUA and ACUUU) and V1 protected sequences (cyan) are shown. (E) Ribonucleoprotein immunoprecipitation (RNP-IP) of the RNA aptamer with an anti-β-catenin antibody. HEK293T cells were transfected with the U6-aptamer containing the wild-type (wt, ACUUU) or mutant (mut, GCGCG) sequence at nucleotides 50–54. Normal IgG was used as a control. Supernatant RNA is shown as a control for RNA aptamer expression.