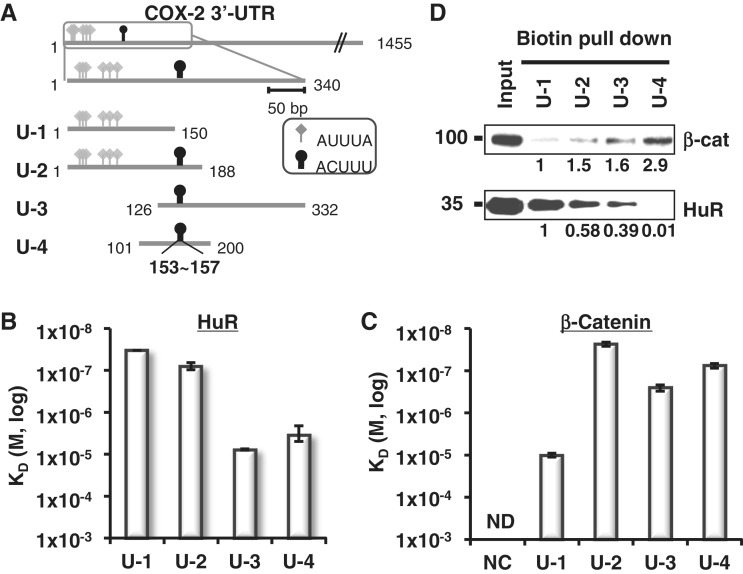
Figure 3.
Mapping of β-catenin and HuR binding on COX-2 3′-UTR. (A) Diagram of COX-2 3′-UTR showing ARE clusters (340 nt, box). Four fragments (U-1 to U-4) were generated spanning ARE clusters. HuR binding elements included six class I/II AREs and are shown with diamond arrowheads (AUUUA). Putative β-catenin binding elements are also shown with oval arrowheads (153–157, ACUUU). Starting and terminating nucleotides are shown. (B) HuR-binding affinity (KD) from SPR analysis with immobilized COX-2 3′-UTR fragments. Three independent SPR experiments were performed and the average KD values of each experiment were evaluated and shown with standard deviations. (C) β-Catenin-binding affinity (KD) of the COX-2 3′-UTR fragments and NC RNA. Three independent SPR experiments were performed and are shown as in C, ND, not detectable. (D) Biotin pull-down analysis of U-1 to U-4 using HT-29 colon cancer cell extracts. β-Catenin and HuR proteins in the RNA-bound pellet fractions were detected by Western blot analysis. Relative fold binding of proteins on U-2, U-3 and U-4 RNA was compared with those of U-1 RNA bound proteins.