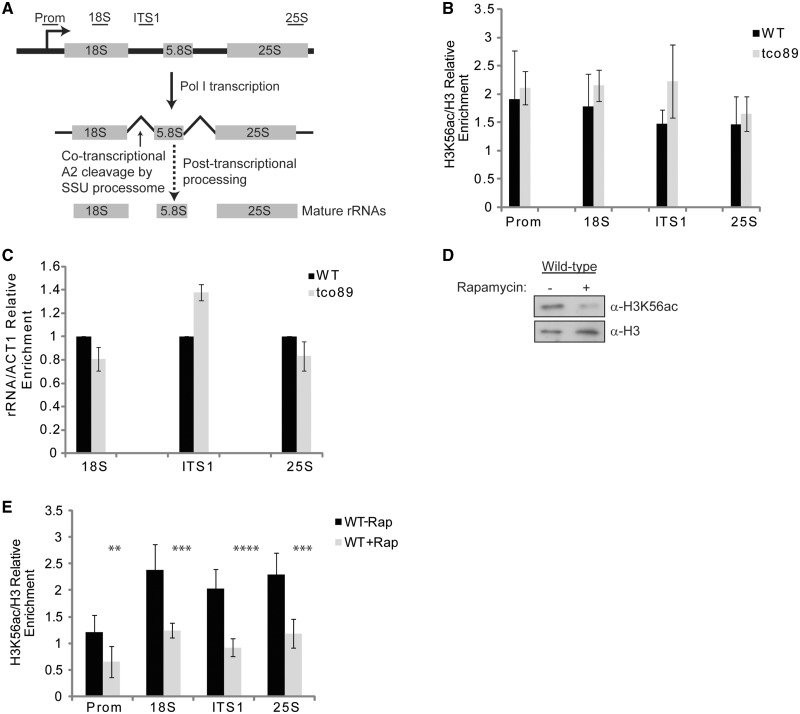
Figure 4.
H3K56ac regulators localize to the rDNA. (A) Simplified schematic of the rDNA locus and the relative locations of primers used in qPCR. Note that the ITS1 specific primers span the A2 site cleaved by the SSU processome during rRNA processing. Detailed description of rRNA processing reactions involved in rRNA maturation are described previously (49). (B) H3K56ac localizes to rDNA but is not decreased in tco89Δ. ChIP using α−H3K56ac or α−H3 antibodies and primers to the promoter, 18S, ITS1 or 25S rDNA sequences. Data are the average and standard deviation of a minimum of two or more independent experiments. (C) tco89Δ does not decrease rRNA levels. Total RNA was prepared from wild-type (WT) and tco89Δ cells grown to log phase and cDNA was synthesized using 1 µg of RNA and random hexamer primers. qPCR was performed with the indicated primer sets, the WT signal was set to 1 and tco89Δ expressed relative to WT The average and standard deviation of two independent experiments are shown. (D) H3K56ac is significantly reduced in WT cells upon rapamycin treatment. WT cells were grown to log phase and either mock treated or treated with an inhibitory (200 nM) concentration of rapamycin for 1 h. WCE were prepared and 30 µg analyzed by SDS–PAGE and α-H3K56ac and α-H3 immunoblotting. (E) Rapamycin mediated inhibition of TORC1 signaling reduces H3K56ac on the rDNA. Experiment was performed as in (D) except samples were processed for α-H3K56ac or α-H3 ChIP. The regions of the rDNA are indicated analyzed are indicated. The data are the average and standard deviation of three or more independent experiments. **P < 0.05; ***P < 0.01; ****P < 0.005.