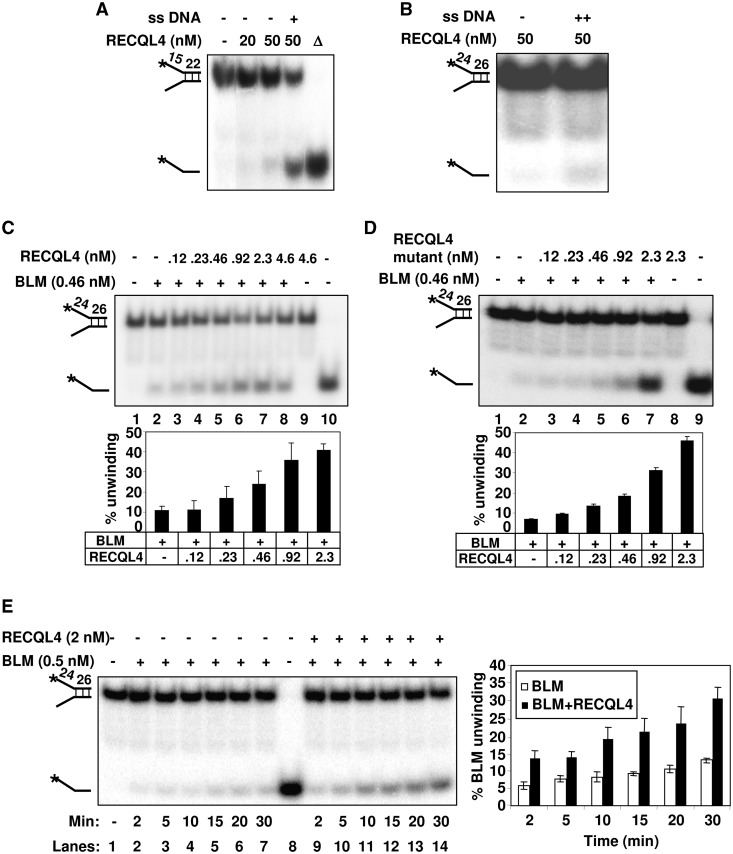
Figure 2.
The helicase activity of BLM is stimulated in the presence of RECQL4. (A) and (B) show the helicase activity of the wild type RECQL4 on DNA fork substrate with 22-mer and 26-mer duplexes in the absence or the presence of 50-fold excess of single-strand complimentary oligonucleotide as indicated. (C) The stimulation of BLM helicase activity in the presence of increasing concentrations of Wt RECQL4 (lanes 3–8). Helicase activities of BLM or RECQL4 alone are shown in lane 2 and 9, respectively. Substrate alone and heat denatured substrate are shown in lanes 1 and 10, respectively. (D) The BLM helicase activity is stimulated in the presence of increasing concentrations of RECQL4 helicase-dead mutant protein (lanes 3–7). Helicase activities of BLM or RECQL4 helicase-dead mutant protein alone are shown in lanes 2 and 8, respectively. The bar graph representing the percent substrate unwinding activity of BLM in the presence of increasing concentrations of RECQL4 Wt or RECQL4 helicase-dead mutant proteins is shown below panels A and B, respectively. (E) The RECQL4 increases the kinetics of BLM unwinding on DNA fork substrate. The helicase reactions were terminated at 2, 5, 10, 15, 20 and 30 min, and analyzed on 8% polyacrylamide gel. The unwinding activity of the BLM protein alone at different time points (as indicated) is shown in lanes 2–7. The unwinding activity of BLM in the presence of Wt RECQL4 protein (1:4 molar ratio) at different time points is shown in lanes 9–14. The bar graph compares the kinetics of substrate unwinding by BLM in the absence or the presence of RECQL4 (1:4 molar ratio) at different time point as indicated.