Abstract
Gene expression is translationally regulated during many cellular and developmental processes. Translation can be modulated by affecting the recruitment of mRNAs to the ribosome, which involves recognition of the 5′ cap structure by the cap-binding protein eIF4E. Drosophila has several genes encoding eIF4E-related proteins, but the biological role of most of them remains unknown. Here, we report that Drosophila eIF4E-3 is required specifically during spermatogenesis. Males lacking eIF4E-3 are sterile, showing defects in meiotic chromosome segregation, cytokinesis, nuclear shaping and individualization. We show that eIF4E-3 physically interacts with both eIF4G and eIF4G-2, the latter being a factor crucial for spermatocyte meiosis. In eIF4E-3 mutant testes, many proteins are present at different levels than in wild type, suggesting widespread effects on translation. Our results imply that eIF4E-3 forms specific eIF4F complexes that are essential for spermatogenesis.
Keywords: Male meiosis, Cytokinesis, Translational control, Fertility
INTRODUCTION
Translational control plays a crucial role during development when transcription is quiescent or when asymmetric localization of proteins needs to be established, such as in early embryogenesis, neurogenesis and oogenesis (Kong and Lasko, 2012; Sonenberg and Hinnebusch, 2007; Thompson et al., 2007). An important example is male gametogenesis, as sperm development relies largely on the translational control of many mRNAs that are translationally silenced in early stages and later differentially activated in space and time (Fuller, 1998; Kleene, 2003; Renkawitz-Pohl et al., 2005; Schäfer et al., 1995).
Most eukaryotic mRNAs are translated in a cap-dependent manner, which requires the recognition of the mRNA 5′ cap structure [m7G(5′)ppp(5′)N, where N is any nucleotide] by the cap-binding protein complex eIF4F. The complex consists of eIF4E, which is the cap-binding subunit; eIF4A, an RNA helicase; and eIF4G, a scaffolding protein that binds poly(A)-binding protein and eIF3. eIF3 also binds the 40S ribosomal subunit. Thus, through eIF4F the mRNA is recruited to the ribosome. The resulting 43S initiation complex then scans along the mRNA 5′ untranslated region (UTR). Upon encountering the start codon, the initiation factors are released, and the 60S ribosomal subunit joins to form an 80S initiation complex (Jackson et al., 2010; Sonenberg and Hinnebusch, 2009).
As translation is mainly controlled at the initiation step, different mechanisms have evolved to regulate eIF4E activity (Hernández et al., 2010; Jackson et al., 2010; Sonenberg and Hinnebusch, 2009). Most eukaryotes have several paralogs of eIF4E (Hernández and Vazquez-Pianzola, 2005; Joshi et al., 2005), but the biological relevance of most of them is unknown. For example, Drosophila has eight eIF4E cognates (Hernández et al., 2005; Hernández et al., 1997; Hernández and Sierra, 1995; Lasko, 2000; Lavoie et al., 1996; Maroto and Sierra, 1989), but the biological functions of only eIF4E-1 (Gong et al., 2004; Graham et al., 2011; Hernández et al., 2005; Hernández et al., 2004; Lachance et al., 2002; Menon et al., 2004; Ottone et al., 2011; Piccioni et al., 2005; Sigrist et al., 2000) and 4E-HP (Cho et al., 2005) have been characterized. Many of the eIF4E paralogs are expressed only in certain types of cells or during distinct developmental processes (Hernández and Vazquez-Pianzola, 2005). This suggests that a ubiquitous eIF4E isoform might carry out global translation initiation, whereas the other isoforms might be involved in more specific processes of translation (Hernández and Vazquez-Pianzola, 2005). Work in C. elegans supports this, as IFE-1, a germline-specific eIF4E, is required for sperm (Amiri et al., 2001; Kawasaki et al., 2011) and oocyte (Henderson et al., 2009) maturation, loss of IFE-2 affects chromosome segregation at meiosis (Song et al., 2010), and IFE-4 is involved in translating only a small set of neural and muscle mRNAs involved in egg laying (Dinkova et al., 2005).
Here, we report that Drosophila eIF4E-3 (which is encoded by CG8023) is a testis-specific protein that controls translation initiation exclusively during male germline development. eIF4E-3 is required for meiotic chromosome segregation and cytokinesis, as well as for later stages of sperm development, including nuclear shaping and individualization. Hence, eIF4E-3 mutants fail to form mature, individual sperm. Our results provide further evidence that alternative forms of eIF4E add complexity to the control of gene expression in eukaryotic development.
MATERIALS AND METHODS
Construction of plasmids
eIF4E cognate cDNAs (Hernández et al., 2005) were subcloned into the vector pET30a (Novagen) without a tag to create expression plasmids pET30-eIF4Es, and into the pOAD vector (Cagney et al., 2000) in-frame with the activator domain sequence of GAL4 to generate plasmids pAD-eIF4Es (‘prey’). eIF4E-3 cDNA was also subcloned into the vector pRSET (Invitrogen) in-frame with the 6×His tag to create expression construct pRSET-4E3. 4E-BP (Thor – FlyBase) cDNA (Miron et al., 2001) and an NcoI fragment of eIF4G-2 encoding amino acids 313-1164 including the eIF4E-binding motif YSIETLR (Baker and Fuller, 2007) were subcloned into the pOBD2 vector (Cagney et al., 2000) in-frame with the DNA-binding domain sequence of GAL4 to create plasmids pBD-4EBP and pBD-eIF4G-2 (313-1164) (‘bait’).
Yeast two-hybrid system
Interactions between bait and prey proteins were detected following a yeast interaction-mating method using Saccharomyces cerevisiae strains PJ69-4A and PJ69-4α (Cagney et al., 2000). Diploid cells containing both bait and prey plasmids were grown in selective media (–Trp, –Leu) and shown as growth control. Protein interactions were detected by replica plating diploid cells onto selective media (–Trp, –Leu, –Ade) or [–Trp, –Leu, –His, + 30 mM 3-amino-1,2,4-triazole (3AT)]. Growth was scored after 4-6 days of growth at 30°C.
Co-immunoprecipitation (co-IP) from testis
Testes were dissected at room temperature from 4- to 8-day-old males in S2 Schneider’s medium supplemented with 10% fetal bovine serum (Gibco-BRL). Medium was removed and 50 testis pairs were ground on ice in 250 μl IP buffer [100 mM KCl, 20 mM HEPES-KOH pH 7.6, 1 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM PMSF, 35 μg/ml RNase A, complete EDTA-free protease inhibitor cocktail (Roche)]. Lysates were spun for 8 minutes at 10,000 rpm (9200 g) at 4°C and the supernatant was precleared for 1 hour at 4°C with 50 μl 50% slurry protein-G–Sepharose (4 Fast Flow, GE Healthcare). Supernatants were then transferred to another tube containing 30 μl 50% slurry protein-G–Sepharose beads with or without primary antibody: 1 μl affinity-purified rabbit anti-eIF4E-1 (#36530) (Lachance et al., 2002); 5 μl (0.28 μg/μl) affinity-purified anti-eIF4E-3 (#968); or 5 μl (0.2 μg/μl) mouse monoclonal anti-HA antibodies (sc-7392, Santa Cruz). The mixture was rotated for 2 hours at 4°C and the beads were then washed four times with 1.5 ml IP buffer. Laemmli sample buffer was added to the beads, which were then boiled for 5 minutes and analyzed by SDS-PAGE and western blot using the ONE-HOUR IP-Western Kit (GenScript).
Generation of antibodies and western blot analyses
Polyclonal affinity-purified anti-eIF4E-3 antibodies #967 and #968 (Biomatik, Ontario, Canada) were raised in rabbit against the peptide ELMSGNEEELQPSLNRVMKNID (amino acids 39-60) of Drosophila eIF4E-3 (Hernández et al., 2005). Recombinant His6×-eIF4E-3 protein was produced by transforming the expression construct pRSET-4E3 into E. coli BL21(DE3) (Novagen), according to the manufacturer’s instructions. The polyclonal antiserum anti-eIF4E-3 #39-3 was raised in rat against His6×-eIF4E-3 protein.
Western blot analyses were performed with the following primary antibodies and working dilutions: affinity-purified anti-eIF4E-3 (#967 and #968), 1:2500; rat affinity-purified anti-eIF4E-3 (#39-3), 1:2000; rabbit affinity-purified anti-eIF4E-1 (#36530) (Lachance et al., 2002), 1:1000; rabbit anti-eIF4G (Zapata et al., 1994), 1:2000; rabbit affinity-purified anti-4E-BP (#1868-3) (Miron et al., 2001), 1:1000; mouse monoclonal anti-HA-HRP (mAb 3F10, Roche), 1:2000; and mouse monoclonal anti-α-tubulin (clone DM 1A, Sigma), 1:5000. Horseradish peroxidase-conjugated anti-rabbit (1:5000) and anti-rat (1:2500) secondary antibodies (GE Healthcare) and the ECL reagent (PerkinElmer) were used to detect primary antibodies.
Immunohistochemistry
Testes were dissected on ice in S2 Schneider’s medium supplemented with 10% fetal bovine serum. Medium was removed and testes were fixed in fixer solution [200 μl of 4% paraformaldehyde in PBST (PBS with 0.2% Tween 20), 600 μl heptane and 20 μl DMSO] for 20 minutes with slow rotation. Fixer was removed and testes were washed three times for 15 minutes each in 1.5 ml PBST followed by 1-2 hours blocking with 1 ml blocking solution (PBST, 0.1% Triton X-100, 1% BSA). Testes were incubated either with phalloidin or with primary antibodies in blocking solution at 4°C overnight, washed for 30 minutes in PBST, blocked for 30 minutes with 500 μl blocking solution containing 1% goat serum, and incubated with secondary antibodies in 500 μl blocking solution containing 8% goat serum overnight at 4°C. At this step, testes were counterstained with DAPI (1 ng/ml) for 20 minutes, washed four times for 20 minutes each with 1.5 ml PBST, and mounted for imaging.
Antibody dilutions for testes staining were as follows: rabbit affinity-purified anti-eIF4E-1 #36530 (Lachance et al., 2002), 1:1000; rat affinity-purified anti-eIF4E-3 #39-3, 1:100; affinity-purified anti-eIF4E-3 #967, 1:300; rabbit anti-eIF4G (Zapata et al., 1994), 1:1000; mouse anti-HA antibodies (sc-7392, Santa Cruz, 0.2 μg/μl), 1:500. Goat fluorescent Alexa Fluor 488- or 546-conjugated secondary antibodies (Molecular Probes) were used at 1:500 to detect primary antibodies. Actin was detected with phalloidin TRITC-546 (Sigma) at 0.2 μM. Imaging was performed with a Leica DM6000B fluorescence microscope.
Squashed preparations of Drosophila testes
Adult testes from 1- to 3-day-old males were dissected in testis isolation buffer (TIB) (Casal et al., 1990) with Hoechst 33342 (1:2000 or 8.3 μg/ml; Sigma) to stain DNA. Squashed preparations of live Drosophila male germ cells were made as previously described (Fabian et al., 2010). The preparations were examined on an epifluorescence microscope (Zeiss Axioplan 2) using Axiovision software (Zeiss). Images were adjusted similarly for brightness, contrast and pseudocolor with Adobe Photoshop.
Crude extract preparation
Total lysates of Drosophila tissues or staged animals were prepared by disrupting on ice 1 g tissue per 1 ml buffer [8 M urea, 20 mM HEPES-KOH pH 7.5, 100 mM KCl, 2 mM EDTA, 0.5% Triton X-100, 1 mM DTT, 2 mM PMSF, complete EDTA-free protease inhibitor cocktail (Roche)], followed by centrifugation at 10,000 rpm (9200 g) for 10 minutes at 4°C. Supernatants were recovered and stored at –80°C. Protein concentration was quantified with the Protein Assay Kit (Bio-Rad).
Purification of untagged recombinant eIF4E and 4E-HP proteins
pET30-eIF4E plasmids were transformed into E. coli BL21(DE3) or Rosetta 2(DE3) (Novagen). Bacteria were grown in LB medium to OD600nm=1 and induced with 0.5 mM IPTG for 3 hours at 37°C. Cells were harvested, resuspended in lysis buffer (20 mM HEPES-KOH pH 7.5, 100 mM KCl, 1 mM EDTA, 2 mM DTT, 10% glycerol) and disrupted by sonication. After centrifugation of the lysate (30,000 g for 30 minutes) the supernatant was removed and the pellet was washed three times with wash buffer (20 mM HEPES-KOH pH 7.2, 1 M guanidine hydrochloride, 2 mM DTT, 10% glycerol). The inclusion bodies were dissolved in 50 mM HEPES-KOH (pH 7.2), 6 M guanidine hydrochloride, 10% glycerol, 2 mM DTT, and cell debris was removed by centrifugation (43,000 g for 30 minutes). Proteins (diluted to below 0.1 mg/ml) were refolded by one-step dialysis against 50 mM HEPES-KOH (pH 7.2), 100 mM KCl (or 50 mM KCl in the case of eIF4E-3), 1.0 mM EDTA, 2 mM DTT, and purified by ion exchange chromatography on a HiTrap SP column (GE Healthcare).
Cap-affinity chromatography
Untagged recombinant protein at 0.1 μM in 500 μl cap-binding buffer (CBB; 100 mM KCl, 20 mM HEPES-KOH pH 7.6, 7 mM β-mercaptoethanol, 0.2 mM EDTA, 10% glycerol, 0.1% Triton X-100, 50 mM β-glycerol phosphate, 0.1 mM GTP, complete EDTA-free protease inhibitor cocktail) was incubated for 1 hour with 50 μl m7GTP-Sepharose 4B (GE Healthcare) at 4°C. The supernatant was recovered (unbound fraction) and the resin was washed five times with 1 ml CBB containing 0.2 mM GTP. A final wash of 20 μl was kept for gel analysis (flow-through). Proteins were further eluted with 20 μl CBB containing 0.4 mM free m7G(5′)ppp(5′)G (New England Biolabs). Proteins from the unbound fraction were precipitated with 40% trichloroacetic acid (TCA) for 30 minutes on ice, centrifuged at 13,000 rpm (15,500 g) for 10 minutes at 4°C, further washed with 1 ml cold acetone, centrifuged 13,000 rpm for 10 minutes at 4°C, air-dried and resuspended in sample buffer. Proteins were subjected to SDS-PAGE and further western blot analyses.
Metabolic labeling with [35S]methionine
Pairs of testes were dissected from 3- to 6-day-old adult males in S2 Schneider’s medium containing L-glutamine supplemented with all amino acids except methionine (Clontech). Twenty pairs of testes were labeled in 400 μl of the same medium containing 80 μCi/ml [35S]Met/Cys mixture (Amersham) for 1.5 hours at 23°C. Testes were washed with Schneider’s medium and lysed in 100 μl RIPA buffer without SDS, and spun at 10,000 rpm (9200 g) for 8 minutes; 20 μl of the supernatant was used to measure incorporation of radioactivity.
Two-dimensional difference gel electrophoresis (2D-DiGE)
eIF4E-3 mutant or wild-type males were dissected in PBS, testes were recovered, frozen in liquid nitrogen and stored at –80°C. Testes were then homogenized in lysis buffer, centrifuged at 10,000 g for 10 minutes at 4°C, and supernatants collected. Protein concentrations were measured using the BioRad kit, and adjusted to 5 mg/ml. Four independent extracts, each prepared from 100 testes pairs, were subjected individually to 2D-DiGE according to the manufacturer’s protocols (GE Healthcare). Briefly, 50 μg of each extract was labeled with either the Cy3 or Cy5 version of the DiGE CyDyes in a randomized fashion. A mixture of equal amounts of protein from each sample was labeled with Cy2 as an internal standard, and 50 μg of that standard mixture was combined with each wild type and mutant extract pair and applied by cup loading to a 24-cm, pH 3-10 gradient strip (Immobiline DryStrip, GE Healthcare) for isoelectric focusing on an IPGphor II instrument (GE Healthcare). The second dimension was run in parallel on 12.5% SDS-PAGE gels in a DALTsix apparatus (GE Healthcare). Gels were imaged immediately after electrophoresis on a Typhoon Trio Plus imager (GE Healthcare). Spot finding, normalization, matching and statistical analysis were carried out using the Decyder software package (version 7, GE Healthcare).
Genomic DNA preparation and PCR amplification
Genomic DNA was isolated from 15 males and PCR amplification performed as previously described (Bellen et al., 2004). Primers (BioCorp) were (5′-3′): a, CGCAACGAGATGGTCACATGACTTTTCCAA; b, CAATCATATCGCTGTCTCACTCAGACTCAATAC; and c, ACTGCTCTACGCAATACTTTATTCATGGTA.
Drosophila strains and fly work
Oregon-R served as wild-type flies. y1 w1118; P{w+mC=lacW}l(3)L0139L0139/TM3, Ser1 (Bier et al., 1989); w1118; Df(3L)BSC732/TM6C, Sb1 cu1; w1118; Df(3L)Exel6112, P{w+mC=XP-U}/TM6B, Tb1; w1118; Df(3L)ED4408, P{w+mW.Scer\FRT.hs3=3′.RS5+3.3′}/TM6C, cu1 Sb1; w1118; Df(3L)BSC376/TM6C, Sb1 cu1 (Parks et al., 2004; Ryder et al., 2004) and nos-Gal4::VP16 fly stocks, were obtained from the Bloomington Drosophila Stock Center. UAS-eIF4E-3RNAi (w1118; P{GD10616}v34210) and P{w+mC=UAS-Dcr-2.D}1, w1118 (Dietzl et al., 2007) flies were obtained from the Vienna Drosophila RNAi Center. Transgenic w1118; P{w+mC=HA-eIF4G-2} flies (Baker and Fuller, 2007) were obtained from C. Baker and M. Fuller.
Fertility tests were performed at 25°C on standard Drosophila culture media. In each vial, five males or females of a given genotype were mated to 15 wild-type females or five males, respectively, for 48 hours. Flies were then transferred to another vial to lay eggs for 48 hours and then discarded. Five to seven vials per genotype were scored.
RESULTS
eIF4E-3 expression is restricted to primary spermatocytes and early differentiating spermatids
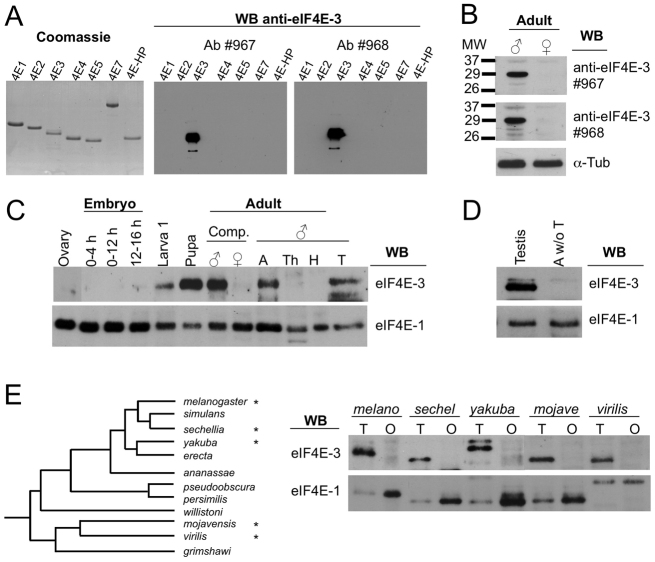
To assess eIF4E-3 expression during development, we raised two rabbit polyclonal antisera against a peptide located at the N-terminus of eIF4E-3, the sequence of which is not conserved in other eIF4Es. These antisera recognized full-length recombinant eIF4E-3 and showed no cross-reactivity to other eIF4Es (Fig. 1A). Both antisera detected eIF4E-3 in male but not female adults (Fig. 1B). Further analysis showed that eIF4E-3 is expressed in larvae and pupae, and only in the abdomen and testes of the adult male (Fig. 1C, lanes 5-7, 9 and 12). eIF4E-3 was not detected in the abdomen of flies without testes (Fig. 1D). The restricted expression of eIF4E-3 in testes was confirmed using new antisera raised in rats (supplementary material Fig. S1). These results demonstrate that eIF4E-3 is a testis-specific protein, and are consistent with transcriptome analysis that found eIF4E-3 mRNA to be highly expressed in testes (Graveley et al., 2011). By contrast, eIF4E-1 is expressed in all the tissues analyzed (Fig. 1C). Sexually dimorphic expression of eIF4E-3 in gonads is conserved across the genus Drosophila (Fig. 1E).
Fig. 1.
eIF4E-3 is a testis-specific protein. (A) Generation of two specific antibodies (#967 and #968) against Drosophila eIF4E-3. (Left) Recombinant Drosophila eIF4Es and 4E-HP proteins (2 μg per lane) were resolved by SDS-PAGE and stained with Coomassie Blue. (Center and right) Western blotting of recombinant eIF4Es (0.2 μg/lane) shows that antibodies #967 and #968 specifically recognize eIF4E-3. (B) Western blot (WB) of total extract (20 μg per lane) from whole wild-type males or females with the indicated antibodies. α-Tubulin provided a loading control. (C,D) Developmental expression of eIF4E-3 and eIF4E-1 as analyzed by western blotting. Total protein extracts (20 μg per lane) prepared from the indicated developmental stages were probed. In all cases western blotting with the respective preimmune sera did not recognize any band (data not shown). (C) Lane 1, ovary; lanes 2-4, embryo lysates from the indicated ages; lane 5, first instar larvae stage; lane 6, pupae; lane 7, whole adult males; lane 8, whole adult females; lanes 9-12, adult males: A, abdomen; Th, thorax; H, head; T, testes. (D) Western blots of adult testes and abdomen without testes (A w/o T). (E) Expression of eIF4Es in gonads across Drosophila species. (Left) Phylogenetic tree of the 12 sequenced species (Clark et al., 2007); species that were analyzed are marked with asterisks. (Right) Western blotting (20 μg/lane) of total lysates form adult testis (T) and ovary (O) from D. melanogaster (melano), D. sechellia (sechel), D. yakuba (yakuba), D. mojavensis (mojave), and D. virilis (virilis) were probed for the indicated proteins. Note that the sexually dimorphic expression of eIF4E-3 (restricted to testis) and eIF4E-1 (in both tissues, but more abundant in the ovary) is conserved across the genus Drosophila.
Next, we studied the expression pattern of eIF4E-3 within the testis. Immunostaining revealed that eIF4E-3 is expressed in the cytoplasm of germline cells from the primary spermatocyte stage to the early differentiating spermatid stage, immediately prior to the onset of spermiogenesis (Fig. 2A). The same expression pattern was found using any of the rabbit and rat antibodies, and no signal was detected in control testes from mutants lacking eIF4E-3 (see below; Fig. 2B), indicating that the staining is specific. This expression pattern matches that reported for eIF4E-3 mRNA as detected by in situ hybridization (Zhao et al., 2010).
Fig. 2.
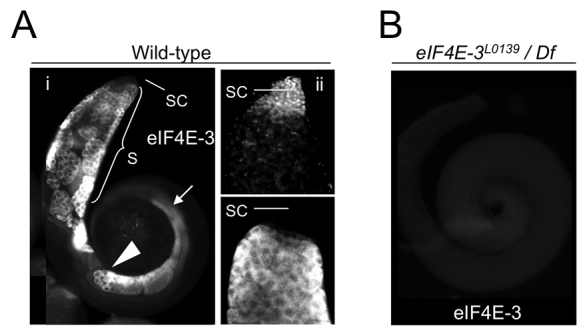
eIF4E-3 is expressed in primary spermatocytes to early differentiating spermatids. Immunostaining of (A) wild-type or (B) eIF4E-3L0139/Df(3L)BSC732 (see Fig. 3) testes with anti-eIF4E-3 antibodies. (Ai) eIF4E-3 is cytoplasmic and expressed in spermatocytes (S), round spermatids (arrowhead) and early differentiating spermatids (arrow), but is absent from the tip of the testis where the stem cells (SC) are located. (Aii) Apical part of a testis showing the tip. Top, DAPI; bottom, eIF4E-3. (B) No signal was detected in eIF4E-3 loss-of-function testes.
eIF4E-3 is essential for male fertility and sperm maturation
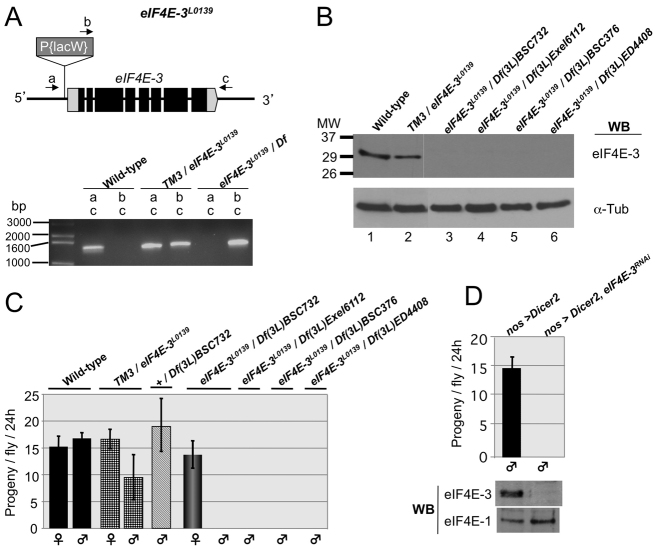
eIF4E-3 is a single-copy gene that encodes a single polypeptide of 244 amino acids that is 59% identical to eIF4E-1 over much of its length (amino acids 66-244). It contains all conserved residues that are needed for cap binding. In predicted eIF4E-3 proteins from many Drosophila species, residues involved in eIF4G/4E-BP binding differ from their eIF4E-1 counterparts in two positions, namely Trp103Phe and Lys160His (Hernández et al., 2005; Lasko, 2000; Tettweiler et al., 2012). To study eIF4E-3 gene function, we used the homozygous-lethal eIF4E-3L0139 mutant, a P-element allele with the insertion 14 nucleotides upstream of the 5′ end of the predicted eIF4E-3 transcription start site (Bier et al., 1989) (Fig. 3A). When eIF4E-3L0139 is in trans with any of four chromosome deficiencies, Df(3L)BSC732, Df(3L)Exel6112, Df(3L)ED4408 or Df(3L)BSC376 (Parks et al., 2004; Ryder et al., 2004), which span the eIF4E-3 gene, eIF4E-3 protein was undetectable by western blotting (Fig. 3B). Flies lacking eIF4E-3 are viable and females are fertile, but males are completely sterile (Fig. 3C). To further demonstrate that eIF4E-3 loss-of-function results in male sterility, we performed eIF4E-3 RNA interference (RNAi) in testes by expressing specific eIF4E-3 hairpin fragments (Dietzl et al., 2007) using the germline-specific nos-Gal4::VP16 promoter. Western blot analysis showed that RNAi expression produced a greater than 95% reduction in eIF4E-3 protein in testes of eIF4E-3RNAi flies, and this led to complete male sterility (Fig. 3D). These complementary experiments confirmed that germline expression of eIF4E-3 is essential for male fertility and rule out any contribution of unidentified second-site mutations to the phenotype.
Fig. 3.
Loss of eIF4E-3 results in male sterility. (A) (Top) The eIF4E-3L0139 gene. Boxes represent exons. Black and gray shading represent coding and untranslated regions, respectively. The P-element P{LacW}L0139 (Bier et al., 1989) inserted within the 5′ untranslated region of eIF4E-3 (CG8023) and the primers (arrows a, b and c) used to verify the P-element insertion are depicted. (Bottom) Agarose gel showing DNA fragments amplified from flies of the indicated genotypes using combinations of primers a, b or c, as indicated. The deficiency used in combination with eIF4E-3L0139 is Df(3L)BSC732. (B) Western blotting of total testis extracts shows that eIF4E-3 is not produced in eIF4E-3L0139 flies when in trans with deficiencies spanning the eIF4E-3 gene (lanes 3-6). (C) Fertility tests of animals of the indicated sex and genotypes. eIF4E-3 loss-of-function flies are completely male sterile. (D) Fertility test of nos-Gal4>UAS-Dicer2 (control) and nos-Gal4>UAS-Dicer2;UAS-eIF4E-3RNAi males shows that RNAi-driven germline-specific knockdown of eIF4E-3 leads to complete sterility. Error bars indicate s.d.
We next studied the cause of sterility in eIF4E-3 mutant males. Before meiosis, cysts of 16 clonally related primary spermatocytes undergo a period of growth and gene expression lasting ∼90 hours. In spermatocyte nuclei, chromatin is concentrated in three distinct regions corresponding to the major bivalents. Each spermatocyte then undergoes meiosis to generate four haploid spermatids, resulting in 64 spermatids per cyst. Early spermatids are round and their chromatin is present throughout the nucleus (Fig. 4A,A′). Each spermatid nucleus is accompanied by a specialized mitochondrial derivative that consists of a pair of giant fused mitochondria wrapped around each other like an onion. During spermatid elongation, the two giant mitochondria unfurl and elongate along the flagellar axoneme (Fig. 4D). As the spermatids elongate, the nuclear chromatin reorganizes and continuously condenses while the nucleus elongates and reduces its volume, resulting in a slim, needle-like sperm head with highly compacted chromatin (Fig. 4F). Then, a cone-like structure composed of F-actin forms within each spermatid. These associate to produce the individualization complex (IC), which assembles around each nucleus in the cyst. The 64 F-actin cones in the cyst move caudally along the spermatid tails, simultaneously expelling excess cytoplasm. Finally, each spermatid is wrapped into an individual membrane, coiled to the base of the testis, and transferred to the seminal vesicle as a mature spermatozoon (Fig. 4H) (Fuller, 1998; Renkawitz-Pohl et al., 2005).
Fig. 4.
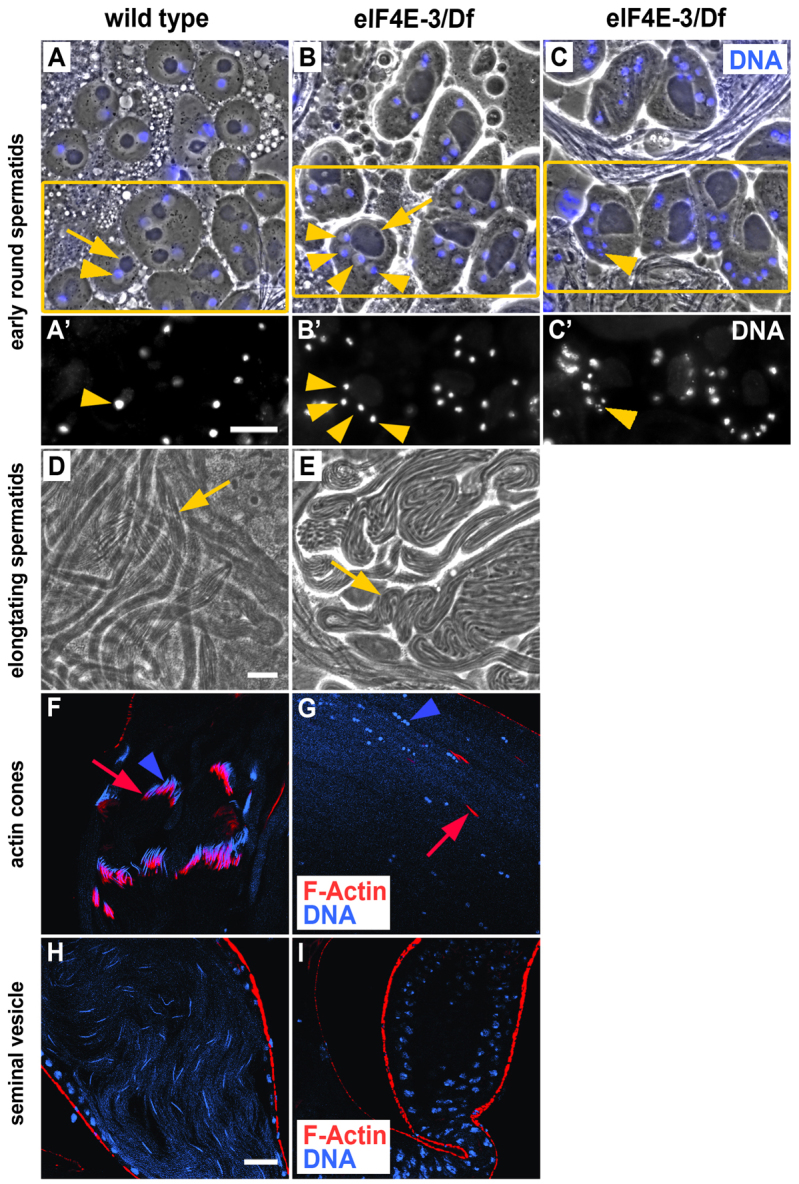
eIF4E-3 is necessary for meiotic cytokinesis and chromosome segregation. (A-C′) Phase-contrast (A-C) and corresponding fluorescence (A′-C′) images of wild-type (A,A′) and eIF4E-3L0139/Df(3L)BSC732 (eIF4E-3) (B-C′) early round spermatids stained for DNA (blue, A-C). Mitochondrial derivatives (arrows) and nuclei (arrowheads) are indicated. Boxes (A-C) indicate regions shown in A′-C′. (D,E) Phase-contrast micrographs of elongating spermatid cysts from wild-type (D) and eIF4E-3L0139/Df(3L)BSC732 (E) testes. Representative cysts are indicated (arrows). Note the thicker, more irregular mitochondrial derivatives (phase-dark linear structures) and fewer elongated cysts in eIF4E-3 as compared with wild type. (F-I) Confocal fluorescence micrographs of wild-type (F,H) and eIF4E-3L0139/Df(3L)BSC732 (G,I) testes stained for F-actin (RITC:phalloidin, red) and DNA (DAPI, blue). (F,G) Elongated spermatid cysts showing mature needle-shaped nuclei and individualization complexes in wild type, and small, condensed nuclei and scattered actin cones in eIF4E-3 mutant. Nuclei, blue arrowheads; actin cones, red arrows. (H,I) Seminal vesicles showing the characteristic needle-like nuclei of mature sperm in wild type and absence of mature sperm in the eIF4E-3L0139/Df(3L)BSC732 mutant, which also shows smaller seminal vesicles. Scale bars: 20 μm.
Phase-contrast imaging of spermatids in eIF4E-3L0139/Df(3L)BSC732 testes revealed defects in meiotic cytokinesis and chromosome segregation. Early round spermatids contained multiple nuclei accompanied by enlarged mitochondrial derivatives (Fig. 4B,B′). In addition, many of these multinucleate spermatids had micronuclei (Fig. 4C,C′). Although eIF4E-3 mutants form elongated spermatid cysts (Fig. 4E), DNA staining revealed defects in the process of nuclear shaping that occurs during spermatid differentiation. The nuclei of eIF4E-3 mutant spermatids remained relatively round, and neither late canoe stage nor the characteristic needle-like nuclei of mature sperm were observed (Fig. 4G). Phalloidin staining, which marks IC F-actin cones, showed that testes lacking eIF4E-3 did not form the IC (compare Fig. 4G with 4F), resulting in complete disruption of the individualization process and an absence of mature sperm (Fig. 4I). In addition, the seminal vesicles were consistently smaller and the basal region was less dense than in wild-type testes (not shown). These phenotypes were completely penetrant in eIF4E-3L0139/Df(3L)BSC732 testes. We conclude that, at a minimum, eIF4E-3 plays a key role in chromosome segregation and cytokinesis during male meiosis. The nuclear shaping and individualization defects we observed might be indirect consequences of prior meiotic defects.
eIF4E-3 promotes translation
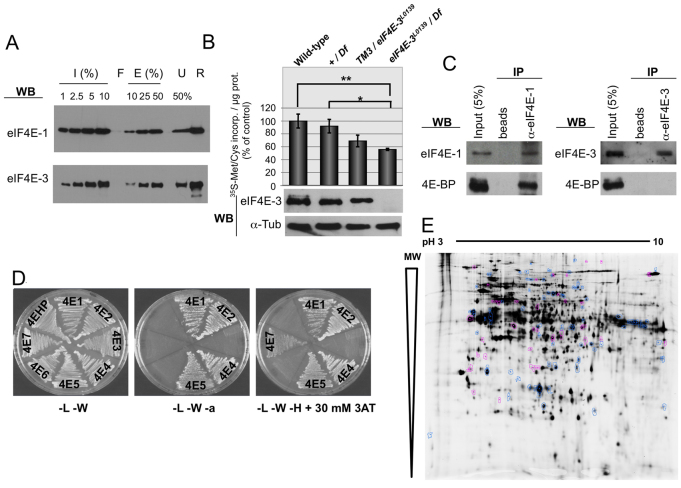
We next investigated whether eIF4E-3 promotes translation in testes. To measure cap binding, we performed pull-down assays on m7GTP-Sepharose using untagged recombinant proteins and further detection by western blotting. Both eIF4E-1 and eIF4E-3 bound the m7GTP-Sepharose resin and were specifically eluted with free cap structure analog (Fig. 5A). Using ImageJ (NIH) software, we quantitated input protein at four different concentrations to produce a densitometry curve, and used it to measure the amount of eluted protein, quantitated in the same manner, at two different concentrations. When calculated in this manner, the proportion of cap-bound eIF4E-1 and cap-bound eIF4E-3 ranged from 10-24% and 9-14%, respectively, over three experiments. We then analyzed the effect of eIF4E-3 loss-of-function on translation in testes. Metabolic labeling of intact testes with a mixture of [35S]Met/Cys ex vivo was performed. We observed a slight reduction in [35S]Met/Cys incorporation in heterozygous TM3/eIF4E-3L0139 as compared with wild-type testes, and a more substantial reduction was observed in mutant testes lacking eIF4E-3, where translation was reduced to ∼50% of wild-type levels (Fig. 5B).
Fig. 5.
eIF4E-3 promotes translation in testis. (A) m7GTP-Sepharose pull-down. Recombinant eIF4E-1 and eIF4E-3 proteins were subjected to m7GTP-Sepharose chromatography. Input (I), flow-through (F), eluted (E) and unbound (U) fractions, as well as the resin (R) after elution, were subjected to SDS-PAGE and western blotting to detect the indicated proteins. (B) Translation in testis as measured by [35S]Met/Cys incorporation ex vivo. (Top) [35S]Met/Cys incorporation of testes of the indicated genotypes. The results of three independent experiments are shown with s.d. Extremely significant (**, P<0.0005) and significant (*, P=0.0186) differences (Student’s t-test, GraphPad) were found between eIF4E-3 loss-of-function [eIF4E-3L0139/Df(3L)BSC732] and wild-type and +/Df(3L)BSC732 values, respectively. (Bottom) The presence of eIF4E-3 and α-tubulin (loading control) in the indicated testes was analyzed by western blotting, probing 20 μg/lane total protein extracts. (C) Co-immunoprecipitation (co-IP) experiments showing that 4E-BP physically interacts with eIF4E-1 but not with eIF4E-3 in testis. Total testis extracts were used to conduct immunoprecipitations using either beads alone or beads plus the indicated antibodies in the presence of RNase A, and interactions were detected by immunoblotting. (D) 4E-BP interacts with eIF4E-1, -2, -4 and -5, but not with eIF4E-3, in the yeast two-hybrid system. 4E-HP and eIF4E-6 were used as negative controls. L, leucine; W, tryptophan; a, adenine; H, histidine; 3AT, 3-amino-1,2,4-triazole. (E) Comparison of testis protein extracts from wild-type and eIF4E-3 mutant flies by 2D-DiGE. Out of ∼2000 protein spots, 120 differ by at least 1.5-fold between wild type and mutant (t-test, P≤0.01). A mixture of wild-type and mutant proteins was run as an internal standard on one of the gels; spots that exhibit an increase in the mutant (38/120) are indicated in magenta, spots that are decreased (82/120) are shown in blue.
We next analyzed whether eIF4E-3 is regulated in testes by the translational repressor 4E-BP by co-immunoprecipitation (co-IP) from total testis extracts in the presence of RNase. Endogenous 4E-BP was associated with eIF4E-1, but not with eIF4E-3 (Fig. 5C). To further analyze interactions between eIF4Es and 4E-BP, we performed yeast two-hybrid experiments. As expected, eIF4E-1, -2, -4 and -5, but not the two negative controls eIF4E-6 and 4E-HP (Hernández et al., 2005), interacted with 4E-BP (Fig. 5D). In addition, eIF4E-3 did not interact with 4E-BP. These results demonstrate that eIF4E-3 promotes translation in testes and suggest that, unusually among eIF4Es, it is not regulated by 4E-BP.
To determine whether eIF4E-3 is highly specific with respect to the mRNAs that it targets, we used 2D-DiGE to compare the relative abundances of ∼2000 proteins in wild-type and eIF4E-3 mutant testes. We identified 120 protein spots (∼6% of the total) with intensities that differed by an average of at least 1.5-fold (P<0.01) between the wild-type and mutant extracts and that changed in the same direction in at least three of the four replicate gels analyzed (Fig. 5E). Consistent with eIF4E-3 acting as a promoter of translation, 82 (68%) of the proteins that changed were reduced in the mutant as compared with wild type; however, 38 of the proteins were increased. We speculate that proteins that increase in level in the mutant might do so if the expression of other eIF4Es were increased when that of eIF4E-3 is reduced or because they are regulated by repressors that are themselves dependent on eIF4E-3 for expression. Overall, these results suggest that eIF4E-3 interacts with a substantial set of mRNAs and thus has a relatively broad role in translation in testes.
From the 2D gels, we recovered 25 and 15 of the spots that were reduced and increased in level, respectively, in eIF4E-3 testes and identified them by liquid chromatography-mass spectrometry (LC-MS). In some cases, more than one spot corresponded to the same protein and the different spots changed their intensity in opposite directions in the eIF4E-3 mutant, suggesting that post-translational modifications were affected. In other cases, the isolated spot contained substantial levels of more than one protein. These were not investigated further. Of 17 clearly identified proteins with reduced levels in eIF4E-3 testes, 14 clustered into the Gene Ontology (GO) term ‘metabolic process’ (GO ID 8152), with 12 of these in ‘cellular metabolic process’ (GO ID 44237) and ten in ‘primary metabolic process’ (GO ID 44238) (supplementary material Table S1). The seven clearly identified proteins that increased in eIF4E-3 testes were more diverse, but four were identified with the GO term ‘small molecule metabolic process’ (GO ID 442). These results indicate that eIF4E-3 functions, at least in part, by regulating metabolic activity.
Another, alternative initiation factor, eIF4G-2, is also a crucial translational regulator during Drosophila meiosis and spermatid differentiation (Baker and Fuller, 2007; Franklin-Dumont et al., 2007). eIF4G-2 mutant males form numerous spermatocytes but these fail to proceed through meiotic divisions, arresting with partially condensed chromosomes prior to establishing the meiotic spindle; thus, elongated spermatids and mature sperm are not found. eIF4G-2 mRNA and protein are expressed in germline cells throughout spermatogenesis, whereas eIF4G mRNA is expressed only in mitotically active cells and early spermatocytes (Baker and Fuller, 2007; Franklin-Dumont et al., 2007). We therefore examined whether eIF4E-3 associates with both eIF4G isoforms. In co-IP experiments using total testes extracts, eIF4E-1 and eIF4E-3 associated with both eIF4G and eIF4G-2 (Fig. 6A). As it was previously shown that eIF4G interacts with both eIF4E-1 and eIF4E-3 in the yeast two-hybrid system (Hernández et al., 2005), we next performed yeast two-hybrid experiments using eIF4G-2 and eIF4Es. eIF4G-2 interacted with both eIF4E-1 and eIF4E-3, but not with the empty vector used as a negative control (Fig. 6B). These results indicate that several eIF4F complexes are present in testis.
Fig. 6.
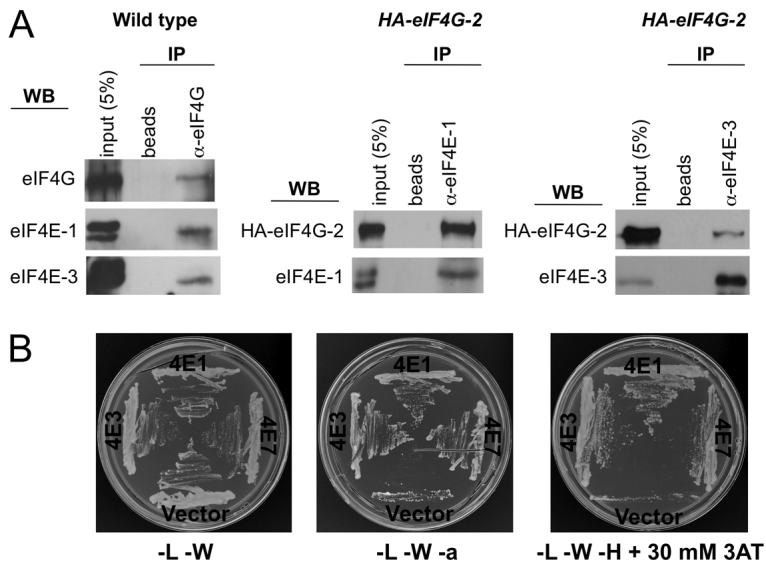
Formation of different eIF4F complexes in testis. (A) co-IP experiments showing that eIF4G (left) and HA-eIF4G-2 (middle and right) physically interact with eIF4E-1 and eIF4E-3 in testis. Total extracts of testes from wild-type or HA-eIF4G-2 flies were used to conduct immunoprecipitations using either beads alone or beads plus the indicated antibodies, and interactions were detected by immunoblotting. (B) eIF4G-2(313-1164) interacts with both eIF4E-1 and eIF4E-3 in the yeast two-hybrid system. Interaction with eIF4E-7 was also observed. Empty vector (pOAD) was used as negative control.
We had noticed that the eIF4E-3 expression pattern (Fig. 2) is similar to that of eIF4G-2 (Baker and Fuller, 2007; Franklin-Dumont et al., 2007). Therefore, we performed co-immunostaining experiments to determine whether these two proteins colocalize within testes. We found that eIF4E-3 and eIF4G-2 colocalize in germline cells from the early spermatocyte stage to the early differentiating spermatid stage, most conspicuously in spermatocytes (Fig. 7A). In agreement with the reported distribution of eIF4G mRNA (Baker and Fuller, 2007; Franklin-Dumont et al., 2007), immunostaining showed that eIF4G protein is enriched in the apical tip of the testis, with a weak and ubiquitous signal in the rest of the testis (Fig. 7B). Unlike eIF4G-2, eIF4G showed only weak colocalization with eIF4E-3 throughout the testis (Fig. 7C). Immunostaining also showed that eIF4E-1 is ubiquitously distributed in the testis and that it colocalizes with eIF4G-2 mainly in spermatocytes (Fig. 7D). Taken together, our results show that eIF4G and eIF4G-2 physically associate and colocalize to different degrees with eIF4E-1 and eIF4E-3, suggesting that they might form four different eIF4F complexes during spermatogenesis.
Fig. 7.
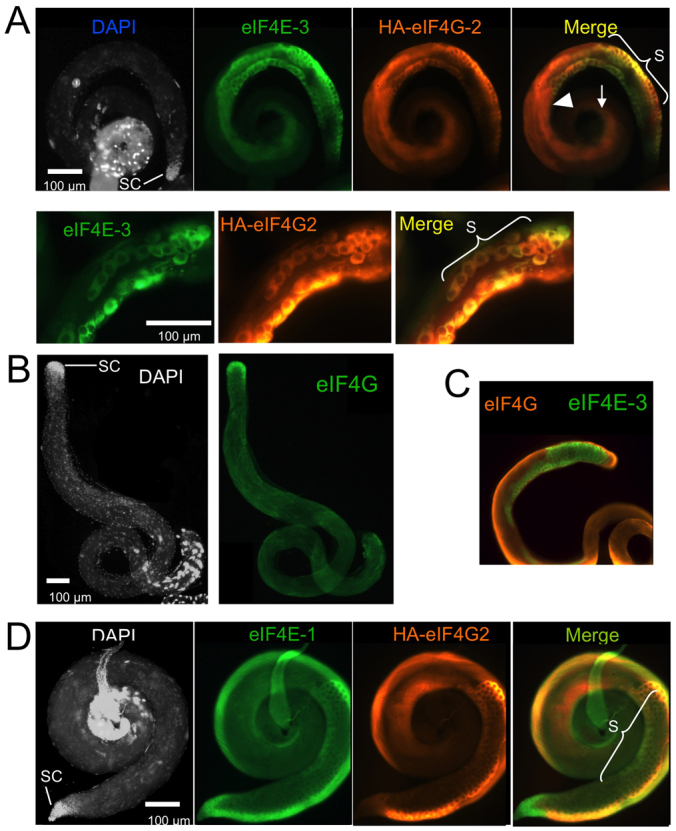
Localization of translation initiation factors in testis. (A) Co-immunostaining on HA-eIF4G-2 testes with anti-eIF4E-3 (green) and anti-HA (which labels HA-eIF4G-2, orange) antibodies. eIF4E-3 and HA-eIF4G-2 colocalize (yellow in merge) in spermatocytes (S), round spermatids (arrowhead) and early differentiating spermatids (arrow), but are absent from the tip of the testis where the stem cells (SC) are located. The higher-magnification images beneath show strong eIF4E-3/HA-eIF4G-2 colocalization in spermatocytes. (B) DAPI staining and immunostaining of wild-type testes with anti-eIF4G antibody. eIF4G shows a ubiquitous distribution with a strong enrichment in the apical tip that contains the stem cells. (C) Co-immunostaining of wild-type testis with anti-eIF4E-3 and anti-eIF4G antibodies. Only weak colocalization is detected. (D) Co-immunostaining of HA-eIF4G-2 testis with anti-eIF4E-1 and anti-HA antibodies. eIF4E-1 (green) is cytoplasmic, ubiquitous, and is enriched in spermatocytes. HA-eIF4G-2 (orange) colocalizes with eIF4E-1 (yellow in merge) mainly in spermatocytes.
DISCUSSION
eIF4E-3 is required for chromosome segregation and cytokinesis at meiosis
It is well established that translational control plays a crucial role in spermatogenesis (Fuller, 1998; Kleene, 2003; Renkawitz-Pohl et al., 2005; Schäfer et al., 1995; White-Cooper and Bausek, 2010). Here, we report that a translation initiation factor, eIF4E-3, is expressed primarily in testes and is required for male fertility. In eIF4E-3 mutant testes, the earliest defects we observed were in meiotic chromosome segregation and cytokinesis. Loss of eIF4E-3 also affected nuclear shaping and sperm individualization. It is possible that eIF4E-3 regulates the translation of one or more factors required for chromosomal segregation and cytokinesis, processes that involve both microtubules and the actin cytoskeleton. Our analysis of protein expression in wild-type and eIF4E-3 mutant testes was thus far limited to abundant proteins, so changes in the levels of regulatory proteins might well have gone undetected. However, the protein that was reduced the most in eIF4E-3 mutant testes was a tektin (CG32820). Tektins are microtubule-stabilizing proteins related to intermediate filament proteins that are essential components of ciliary and flagellar axonemes and of basal bodies and centrioles (Amos, 2008). It is possible that a reduction in the levels of this protein underlies some of the mutant phenotypes we observed.
In this work, we provide further evidence that spermatogenesis in metazoans requires the specialized function of specific eIF4E and eIF4G paralogs and the assembly of non-canonical eIF4F cap-binding complexes, as previously reported for Drosophila eIF4G-2 (Baker and Fuller, 2007; Franklin-Dumont et al., 2007). We extend these previous studies to provide evidence that eIF4E-3 has a broad role in protein synthesis during spermatogenesis. Work in other organisms also supports functions for alternative forms of eIF4E and eIF4G in the germline. It is noteworthy that IFE-2, an alternative eIF4E in C. elegans, has, like Drosophila eIF4E-3, been linked to chromosome segregation at meiosis (Song et al., 2010). This degree of evolutionary conservation suggests an ancient translational mechanism for control of this process in sperm development. Mammals appear to lack a testes-specific eIF4E. However, the mouse repro8 mutation causes an alanine-to-proline substitution in an alternative isoform of eIF4G (eIF4G-3) (Sun et al., 2010). In repro8 mutants, spermatogenesis is completely blocked at meiotic prophase. This occurs as a consequence of a reduction of translation of HSP2A, a spermatogenic cell-specific chaperone protein that is required for activation of CDC2A kinase, which regulates the prophase to metaphase transition. Thus, in invertebrates and mammals, alternative forms of cap-binding complex components are essential for key steps of spermatogenesis. Interestingly, an uncharacterized testes-specific chaperone protein, CG12020, is among the proteins that we identified with reduced levels in Drosophila eIF4E-3 mutant testes (supplementary material Table S1).
Formation of different eIF4F complexes involved in spermatogenesis
The physical interaction between eIF4G-2 and eIF4E-3, their strong cellular colocalization and the ability of eIF4G-2 to bind eIF4E-1 (Fig. 6B) (Baker and Fuller, 2007) suggest that these two alternative isoforms of translation factors form a functional eIF4F complex that is required for translation during spermiogenesis. eIF4E-1 and eIF4G are likely to form an eIF4F complex in testis as well. eIF4E-1–eIF4G-2 and eIF4E-3–eIF4G complexes might also exist as there are overlapping distributions of all four proteins in testes, and these pairs can be co-immunoprecipitated from testes extracts. Thus, our data suggest that the existence of several eIF4 paralogs in an organism allows the combinatorial formation of distinct eIF4F complexes that might selectively translate specific mRNAs. In addition, global analysis of the Drosophila melanogaster transcriptome indicates that expression of three other eIF4Es, namely eIF4E-4, eIF4E-5 and eIF4E-7, is also mostly restricted to testes (Graveley et al., 2011). This implies that there is a far greater diversity of eIF4F complexes in testes than previously realized, raising the possibility of an additional layer of complexity in translational regulation during spermatogenesis. Translational regulation involving the poly(A) tail has been recently linked to late stages of spermatogenesis, as mutations in Gld2, which encodes a cytoplasmic poly(A) polymerase, result in defects in sperm individualization and in chromatin reorganization (Sartain et al., 2011).
Diversification and regulation of eIF4Es in Drosophila
Here, we have provided evidence that eIF4Es have undergone molecular diversification and functional specialization in Drosophila, supporting the hypothesis that, in metazoans, a ubiquitous eIF4E is responsible for global translation initiation, whereas other paralogs function in specific cellular processes (Hernández and Vazquez-Pianzola, 2005). eIF4E-1 and eIF4E-3 genes are present, and eIF4E-3 expression is male-specific, across the genus Drosophila. This suggests that the ancestral gene of eIF4E-1 and eIF4E-3 duplicated and functionally diverged prior to the emergence of the genus Drosophila 65 million years ago (Tamura et al., 2004). One of the important aspects of eIF4E-3 divergence is the mutation of two important residues for eIF4G/4E-BP binding (Trp103 and Lys160), which might explain why eIF4E-3 does not bind 4E-BP.
Phosphorylation at Ser209 regulates mammalian eIF4E activity (Furic et al., 2010; Scheper and Proud, 2002). The counterpart of mammalian Ser209 is present in most eIF4Es from metazoans and Schizosaccharomyces pombe (Joshi et al., 2005), and phosphorylation of eIF4E-1 is crucial for development and cell growth in Drosophila (Lachance et al., 2002). By contrast, Drosophila eIF4E-3 does not possess a Ser209 counterpart, having instead a proline (Pro236) residue that is conserved in eIF4E-3 from all sequenced Drosophila species (Clark et al., 2007; Tettweiler et al., 2012). The counterpart of Ser209 is also absent in eIF4Es from various fungi and plants (Gallie, 2007), and eIF4Es from different eukaryotes are phosphorylated on different residues that are not equivalent to mammalian Ser209 (Nett et al., 2009; Zanchin and McCarthy, 1995). This suggests that phosphorylation might affect eIF4E activity differently in different eukaryotes. Whether Drosophila eIF4E-3 is a phosphoprotein and, if so, what the effects of phosphorylation on its activity might be, remain to be elucidated.
The highly dimorphic nature of sperm and eggs clearly required the evolution of strikingly different genetic regulatory networks in the two cell types (White-Cooper and Bausek, 2010). Indeed, significantly higher rates of evolution in genes with male-biased expression (i.e. male specific: seminal fluid proteins and spermatogenesis) relative to female-specific or somatic counterparts have been found in many taxa, including species across the genus Drosophila and in mammals (Ellegren and Parsch, 2007; Haerty et al., 2007; Larracuente et al., 2008). Moreover, genes with strong tissue-biased expression, including reproduction-associated genes, have higher rates of evolution than ubiquitously expressed genes (Larracuente et al., 2008; Swanson and Vacquier, 2002). Therefore, testis-specific proteins are subject to regulatory mechanisms that are related to the forces driving sperm evolution, and these are different from those regulating their somatic counterparts. The lack of interaction of eIF4E-3 with 4E-BP, the lack of the corresponding phosphorylatable Ser209, and the involvement of alternative eIF4E and eIF4G paralogs in spermatogenesis are likely to be reflections of this.
Supplementary Material
Acknowledgments
We thank Elke Küster-Schöck of the McGill Cellular Imaging and Analysis Network for help with designing and analyzing the DiGE experiment; Isabelle Fernandes and Helen White-Cooper for valuable scientific discussions and technical advice; Misia Kowanda for flies; Catherine Baker and Margaret Fuller for the pMT-HA-eIF4G-2 plasmid and for flies; Stanley Fields for plasmids; José M. Sierra for the anti-eIF4G antibody; Bruno Fonseca and Thomas Sundermeier for useful discussions; and Éric Bonneil and the IRIC proteomic platform at the Université de Montréal for the LC-MS analysis.
Footnotes
Funding
This work was supported by grants from the Canadian Institutes of Health Research [MOP-44050 to P.L. and N.S. and MOP-102546 to J.A.B.] and the National Institutes of Health [R01-HD036631 to P.L.]; V.G. was supported by an EMBO long-term postdoctoral fellowship; and J.Z. was supported by the Polish Ministry of Science and Higher Education [N N301 096339]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.073122/-/DC1
References
- Amiri A., Keiper B. D., Kawasaki I., Fan Y., Kohara Y., Rhoads R. E., Strome S. (2001). An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 128, 3899–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos L. A. (2008). The tektin family of microtubule-stabilizing proteins. Genome Biol. 9, 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. C., Fuller M. T. (2007). Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development 134, 2863–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., Tsang G., Evans-Holm M., Hiesinger P. R., Schulze K. L., Rubin G. M., et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Shepherd S., Lee K., McCall K., Barbel S., Ackerman L., Carretto R., Uemura T., Grell E., et al. (1989). Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 3, 1273–1287 [DOI] [PubMed] [Google Scholar]
- Cagney G., Uetz P., Fields S. (2000). High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 328, 3–14 [DOI] [PubMed] [Google Scholar]
- Casal J., González C., Ripoll P. (1990). Spindles and centrosomes during male meiosis in Drosophila melanogaster. Eur. J. Cell Biol. 51, 38–44 [PubMed] [Google Scholar]
- Cho P. F., Poulin F., Cho-Park Y. A., Cho-Park I. B., Chicoine J. D., Lasko P., Sonenberg N. (2005). A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121, 411–423 [DOI] [PubMed] [Google Scholar]
- Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B., Markow T. A., Kaufman T. C., Kellis M., Gelbart W., Iyer V. N., et al. (2007). Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- Dinkova T. D., Keiper B. D., Korneeva N. L., Aamodt E. J., Rhoads R. E. (2005). Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 25, 100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Parsch J. (2007). The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8, 689–698 [DOI] [PubMed] [Google Scholar]
- Fabian L., Wei H. C., Rollins J., Noguchi T., Blankenship J. T., Bellamkonda K., Polevoy G., Gervais L., Guichet A., Fuller M. T., et al. (2010). Phosphatidylinositol 4,5-bisphosphate directs spermatid cell polarity and exocyst localization in Drosophila. Mol. Biol. Cell 21, 1546–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Dumont T. M., Chatterjee C., Wasserman S. A., Dinardo S. meiosis and differentiation in Drosophila spermatocytes. Development 134, 2851–2861 [DOI] [PubMed] [Google Scholar]
- Fuller M. T. (1998). Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin. Cell Dev. Biol. 9, 433–444 [DOI] [PubMed] [Google Scholar]
- Furic L., Rong L., Larsson O., Koumakpayi I. H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L. A., et al. (2010). eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. USA 107, 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D. R. (2007). Translational control in plants and chloroplasts. In Translational Control in Biology and Medicine (ed. Mathews M. B., Sonenberg N., Hershey J. W. B.), pp. 747–774 New York, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Gong L., Puri M., Unlü M., Young M., Robertson K., Viswanathan S., Krishnaswamy A., Dowd S. R., Minden J. S. (2004). Drosophila ventral furrow morphogenesis: a proteomic analysis. Development 131, 643–656 [DOI] [PubMed] [Google Scholar]
- Graham P. L., Yanowitz J. L., Penn J. K., Deshpande G., Schedl P. (2011). The translation initiation factor eIF4E regulates the sex-specific expression of the master switch gene Sxl in Drosophila melanogaster. PLoS Genet. 7, e1002185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W., Jagadeeshan S., Kulathinal R. J., Wong A., Ravi Ram K., Sirot L. K., Levesque L., Artieri C. G., Wolfner M. F., Civetta A., et al. (2007). Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177, 1321–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson M. A., Cronland E., Dunkelbarger S., Contreras V., Strome S., Keiper B. D. (2009). A germline-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J. Cell Sci. 122, 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G., Sierra J. M. (1995). Translation initiation factor eIF-4E from Drosophila: cDNA sequence and expression of the gene. Biochim. Biophys. Acta 1261, 427–431 [DOI] [PubMed] [Google Scholar]
- Hernández G., Vazquez-Pianzola P. (2005). Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 122, 865–876 [DOI] [PubMed] [Google Scholar]
- Hernández G., Diez del Corral R., Santoyo J., Campuzano S., Sierra J. M. (1997). Localization, structure and expression of the gene for translation initiation factor eIF-4E from Drosophila melanogaster. Mol. Gen. Genet. 253, 624–633 [DOI] [PubMed] [Google Scholar]
- Hernández G., Vázquez-Pianzola P., Sierra J. M., Rivera-Pomar R. (2004). Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA 10, 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G., Altmann M., Sierra J. M., Urlaub H., Diez del Corral R., Schwartz P., Rivera-Pomar R. (2005). Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech. Dev. 122, 529–543 [DOI] [PubMed] [Google Scholar]
- Hernández G., Altmann M., Lasko P. (2010). Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem. Sci. 35, 63–73 [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U., Pestova T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B., Lee K., Maeder D. L., Jagus R. (2005). Phylogenetic analysis of eIF4E-family members. BMC Evol. Biol. 5, 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I., Jeong M. H., Shim Y. H. (2011). Regulation of sperm-specific proteins by IFE-1, a germline-specific homolog of eIF4E, in C. elegans. Mol. Cells 31, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene K. C. (2003). Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet. Genome Res. 103, 217–224 [DOI] [PubMed] [Google Scholar]
- Kong J., Lasko P. (2012). Translational control in cellular and developmental processes. Nat. Rev. Genet. 13, 383–394 [DOI] [PubMed] [Google Scholar]
- Lachance P. E., Miron M., Raught B., Sonenberg N., Lasko P. (2002). Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 22, 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A. M., Sackton T. B., Greenberg A. J., Wong A., Singh N. D., Sturgill D., Zhang Y., Oliver B., Clark A. G. (2008). Evolution of protein-coding genes in Drosophila. Trends Genet. 24, 114–123 [DOI] [PubMed] [Google Scholar]
- Lasko P. (2000). The drosophila melanogaster genome: translation factors and RNA binding proteins. J. Cell Biol. 150, F51–F56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C. A., Lachance P. E. D., Sonenberg N., Lasko P. (1996). Alternatively spliced transcripts from the Drosophila eIF4E gene produce two different Cap-binding proteins. J. Biol. Chem. 271, 16393–16398 [DOI] [PubMed] [Google Scholar]
- Maroto F. G., Sierra J. M. (1989). Purification and characterization of mRNA cap-binding protein from Drosophila melanogaster embryos. Mol. Cell. Biol. 9, 2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K. P., Sanyal S., Habara Y., Sanchez R., Wharton R. P., Ramaswami M., Zinn K. (2004). The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron 44, 663–676 [DOI] [PubMed] [Google Scholar]
- Miron M., Verdú J., Lachance P. E., Birnbaum M. J., Lasko P. F., Sonenberg N. (2001). The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3, 596–601 [DOI] [PubMed] [Google Scholar]
- Nett I. R., Martin D. M., Miranda-Saavedra D., Lamont D., Barber J. D., Mehlert A., Ferguson M. A. (2009). The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteomics 8, 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottone C., Galasso A., Gemei M., Pisa V., Gigliotti S., Piccioni F., Graziani F., Verrotti di Pianella A. (2011). Diminution of eIF4E activity suppresses parkin mutant phenotypes. Gene 470, 12–19 [DOI] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., Huppert K., Tan L. R., Winter C. G., Bogart K. P., Deal J. E., et al. (2004). Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36, 288–292 [DOI] [PubMed] [Google Scholar]
- Piccioni F., Zappavigna V., Verrotti A. C. (2005). A cup full of functions. RNA Biol. 2, 125–128 [DOI] [PubMed] [Google Scholar]
- Renkawitz-Pohl R., Hempel L., Hollman M., Schafer M. A. (2005). Spermatogenesis. In Comprehensive Molecular Insect Science: Reproduction and Development, Vol. 1 (ed. Gilbert L. I., Iatrou K., Gill S. S.), pp. 157–177: Maryland Heights, MO: Elsevier Pergamon; [Google Scholar]
- Ryder E. J., Blows F. M., Ashburner M., Bautista-Llacer R., Coulson D., Drummond J., Webster J., Gubb D., Gunton N. J., Johnson G., et al. (2004). The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167, 797–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain C. V., Cui J., Meisel R. P., Wolfner M. F. (2011). The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster. Development 138, 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M., Nayernia K., Engel W., Schäfer U. (1995). Translational control in spermatogenesis. Dev. Biol. 172, 344–352 [DOI] [PubMed] [Google Scholar]
- Scheper G. C., Proud C. G. (2002). Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 269, 5350–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S. J., Thiel P. R., Reiff D. F., Lachance P. E. D., Lasko P., Schuster C. M. (2000). Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405, 1062–1065 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A. G. (2007). New modes of translational control in development, behavior, and disease. Mol. Cell 28, 721–729 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song A., Labella S., Korneeva N. L., Keiper B. D., Aamodt E. J., Zetka M., Rhoads R. E. (2010). A C. elegans eIF4E-family member upregulates translation at elevated temperatures of mRNAs encoding MSH-5 and other meiotic crossover proteins. J. Cell Sci. 123, 2228–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Palmer K., Handel M. A. (2010). Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development 137, 1699–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W. J., Vacquier V. D. (2002). The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 [DOI] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S. (2004). Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21, 36–44 [DOI] [PubMed] [Google Scholar]
- Tettweiler G., Kowanda M., Lasko P., Sonenberg N., Hernández G. (2012). The distribution of eIF4E family members across Insecta. Comp. Funct. Genomics 2012:960420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B., Wickens M., Kimble J. (2007). Translational control in development. In Translational Control in Biology and Medicine (ed. Mathews M. B., Sonenberg N., Hershey J. W. B.), pp. 507–544: New York, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- White-Cooper H., Bausek N. (2010). Evolution and spermiogenesis. Phil. Trans. R. Soc. 365, 1465–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchin N. I., McCarthy J. E. G. (1995). Characterization of the in vivo phosphorylation sites of the mRNA cap-binding complex proteins eukaryotic initiation factor-4E and p20 in Saccharomyces cerevisiae. J. Biol. Chem. 270, 26505–26510 [DOI] [PubMed] [Google Scholar]
- Zapata J. M., Martínez M. A., Sierra J. M. (1994). Purification and characterization of eukaryotic polypeptide chain initiation factor 4F from Drosophila melanogaster embryos. J. Biol. Chem. 269, 18047–18052 [PubMed] [Google Scholar]
- Zhao J., Klyne G., Benson E., Gudmannsdottir E., White-Cooper H., Shotton D. (2010). FlyTED: the Drosophila testis gene expression database. Nucleic Acids Res. 38, D710–D715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.