Abstract
Sphingosine-1-phosphate (S1P) is a bioactive sphingolipid involved in immunity, inflammation, angiogenesis, and cancer. S1P lyase (SPL) is the essential enzyme responsible for S1P degradation. SPL augments apoptosis and is down-regulated in cancer. SPL generates a S1P chemical gradient that promotes lymphocyte trafficking and as such is being targeted to treat autoimmune diseases. Despite growing interest in SPL as a disease marker, antioncogene, and pharmacological target, no comprehensive characterization of SPL expression in mammalian tissues has been reported. We investigated SPL expression in developing and adult mouse tissues by generating and characterizing a β-galactosidase-SPL reporter mouse combined with immunohistochemistry, immunoblotting, and enzyme assays. SPL was expressed in thymic and splenic stromal cells, splenocytes, Peyer's Patches, colonic lymphoid aggregates, circulating T and B lymphocytes, granulocytes, and monocytes, with lowest expression in thymocytes. SPL was highly expressed within the CNS, including arachnoid lining cells, spinal cord, choroid plexus, trigeminal nerve ganglion, and specific neurons of the olfactory bulb, cerebral cortex, midbrain, hindbrain, and cerebellum. Expression was detected in brown adipose tissue, female gonads, adrenal cortex, bladder epithelium, Harderian and preputial glands, and hair follicles. This unique expression pattern suggests SPL has many undiscovered physiological functions apart from its role in immunity.
Keywords: sphingosine phosphate lyase, colon cancer, development, sphingolipid, signal transduction
Sphingosine-1-phosphate (S1P) is the final metabolic product of sphingolipid degradation. It circulates in the blood and lymph bound to lipoproteins and functions as a ligand for a family of five differentially expressed G protein-coupled receptors, the S1P receptors (S1PRs) (1, 2). Acting through these receptors and the downstream signaling events mediated by them, S1P influences cell migration, survival, and stress responses (3). S1PR signaling is critical for development, as demonstrated by the embryonic lethal phenotype of S1PR1 knockout mice, due to severe hemorrhage associated with impaired vascular maturation (4). S1PR2 knockout mice are deaf due to defects in the stria vascularis (5). Additional studies have demonstrated the importance of S1P transport and signaling in cardiac and brain development, reproduction, and embryonic stem cell survival (6–17). S1P acts through its cognate receptors to control T, B, natural killer, and hematopoietic stem cell trafficking as these cells emerge from bone marrow and thymus and transit peripheral lymphoid organs (18, 19). S1P signaling contributes to the regulation and constitutive activation of key transcription factors that play a central role in inflammation and cancer, including nuclear factor-κB and STAT3 (20, 21). S1P is also a mediator of cardioprotection in response to ischemia and can reduce developmental cell death as well as apoptosis, tissue injury, and infertility in response to ionizing radiation (22–24). Although many effects of S1P signaling are mediated through activation of its receptors, some of its actions, including activation of nuclear factor-κB signaling and regulation of histone deacetylases, appear to be mediated through the receptor-independent, intracellular functions of S1P (21, 25).
S1P is generated via phosphorylation of the sphingoid base sphingosine by sphingosine kinases SphK1 and SphK2 (26). These two enzymes exhibit different biochemical properties, are expressed in different tissues, are located in different subcellular compartments, and manifest distinct biological functions. S1P generated through the actions of sphingosine kinases can be dephosphorylated by the S1P phosphatases SPP1 and SPP2 or through reactions catalyzed by nonspecific lipid phosphate phosphatases, the LPPs (27–29). However, each of these reactions results in regeneration of sphingosine, which is then immediately available to be rephosphorylated. In contrast to the phosphatases, SPL is responsible for catalyzing the irreversible degradation of S1P to hexadecenal and ethanolamine phosphate by cleaving the C2-3 carbon bond in S1P in what constitutes the final step of sphingolipid metabolism (30). As such, SPL functions as the major regulator of S1P levels, and loss of its activity or expression results in significant accumulation of S1P and other sphingolipid intermediates within cells and tissues (31). The enzyme is conserved from yeast to humans, and mutants in various model organisms have revealed phenotypes including abnormalities of growth regulation and carbon metabolism in Saccharomyces cerevisiae, developmental and migration defects in Dictyostelium discoideum, reproductive defects in Caenorhabditis elegans, and defects of reproductive organs and muscles in Drosophila melanogaster (32–36).
Mammalian SPL is encoded by the Sgpl1 gene, found on chromosome 10 of mouse and human genomes (37, 38). Mice homozygous for a Sgpl1 null allele demonstrate negligible SPL expression and enzyme activity in tissues as well as high circulating and tissue levels of S1P, indicating that Sgpl1 is the major or only gene responsible for SPL activity (31, 39). SPL null mutants do not live past 1 month of age postnatally and exhibit a range of phenotypes including skeletal and hematological abnormalities, elevated cytokines and proinflammatory responses, impaired neutrophil and lymphocyte trafficking, elevated serum lipids, increased lipid storage in the liver, and deficient adipose stores (39, 40). These findings collectively implicate SPL in development, lipid homeostasis, and the regulation of innate and adaptive immunity.
Recent studies have implicated SPL as a potential immunomodulatory target in the context of autoimmune disease (41–43). LX2931, now in clinical trials for the treatment of rheumatoid arthritis, acts by inhibiting SPL and disrupting the S1P chemical gradient required for mature T-cell egress from the thymus and peripheral lymphoid organs and thereby preventing immune-mediated tissue damage (43). Studies have also implicated SPL as a potential target for cardioprotection, radioprotection, and recruitment of skeletal muscle stem cells for regeneration of injured muscle (44–46). SPL is down-regulated in colon cancer and other malignancies (47–49). The findings that overexpression of SPL promotes apoptosis in response to DNA damage, whereas knockdown of SPL promotes cell transformation, suggest that SPL silencing contributes to carcinogenesis and that SPL may serve an antioncogenic function by promoting the turnover of DNA-damaged cells (45, 50, 51). Although most of the functions and phenotypes associated with SPL and its disruption are attributed to its regulation of S1P, there is growing evidence that the products of the reaction catalyzed by SPL are important regulators of apoptosis and may have additional biological functions (52, 53).
Despite growing interest in SPL as a disease marker, potential antioncogene, and pharmacological target, no comprehensive characterization of its expression pattern in mammalian tissues has been reported. In the current study, we explored SPL expression in embryonic and adult murine tissues by generating and characterizing a β-galactosidase (β-Gal) SPL reporter mouse, combined with immunohistochemistry (IHC), immunoblotting, and enzyme activity assays. In addition, we have explored SPL expression in the developing mouse embryo. These studies provide a more complete characterization of SPL expression than has been previously reported.
MATERIALS AND METHODS
Generation of SPL β-Gal reporter mice and other animals used
The SPL reporter mouse line was generated during the production of a conditional SPL knockout line. To produce these lines, we generated a sequence-replacement targeting vector designed to insert a gene-trapping cassette containing a lacZ reporter within intron 9-10 of Sgpl1. The gene-trapping vector, which was flanked by Frt sites, consisted of a splice acceptor site (flanked by a loxP and a lox71 site) followed by a promoter-less βgeo cassette. This is the same gene-trapping cassette used by the KOMP: Knock-Out Mouse Project (54). We used the gene-trapping cassette designed to yield an in-frame fusion with exon 9 of Sgpl1. The gene-targeting vector was designed to insert a single loxP site into intron 12-13. The integrity of the completed gene-targeting vector was verified by restriction endonuclease digestion and by DNA sequencing.
The gene-targeting vector was electroporated into strain 129/OlaHsd embryonic stem cells, which were then subjected to selection in G418. G418-resistant embryonic stem cell clones that produced a fusion transcript extending from exon 9 of Sgpl1 to βgeo (indicating the expected recombination event had occurred) were identified by 5′ rapid amplification of 5′ cDNA ends. PCR was used to identify clones that also retained the distant loxP site. One such clone was then injected into C57BL/6 blastocysts to generate chimeric mice. A total of 10 high-percentage male chimeras were obtained, and those mice transmitted the targeted mutation to their offspring. The reporter mice were backcrossed six times and subsequently maintained on the C57BL/6 background.
WT C57BL/6 mice were obtained from Taconic (Oxnard, CA). C57BL/6 mice carrying the Sgpl1 null allele described previously (39) were obtained from Philippe Soriano (Mt. Sinai School of Medicine, New York). Animals were maintained in the Children's Hospital Oakland Research Institute (CHORI) AAALAC Accredited Animal Facility. All experiments were conducted in accordance with CHORI Institutional Animal Care and Use Committee approved protocols.
Whole mount survey of SPL β-Gal reporter expression in adult murine tissues
Organ dissection, fixation, and staining for β-Gal activity in adult male and female reporter mice was conducted essentially as described (55).
Frozen section tissue staining for β-Gal activity
Tissue sections were fixed and stained for β-Gal activity essentially as described (56). Briefly, tissue samples from SPL reporter and wild-type (WT) mice were collected and fixed in LacZ fixative solution containing 0.2% glutaraldehyde, 5 mM EGTA (pH 7.3), and 100 mM MgCl2 in 0.1 M NaPO4 (pH 7.3) for 4 h at room temperature (RT) with a solution change at 2 h. The samples were transferred into 15% sucrose in PBS for 4 h at RT and then into 30% sucrose in PBS overnight at 4°C. Tissues were embedded in OCT, and 10 μm frozen sections were cut. Air-dried sections were washed three times in PBS, transferred to X-Gal staining solution (1 mg/ml of 5-bromo-4-chloro-3-indolyl β-D-galactosidase, 2 mM MgCl2, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 0.01% sodium deoxycholate, and 0.02% Nonidet-P40) overnight at 37°C. The following day, samples were washed in distilled water, counterstained with Nuclear Fast Red, and coverslipped with aqueous mounting media. For each tissue examined, corresponding tissue from a WT mouse lacking the SPL reporter was used as a negative control.
IHC for SPL expression
Formalin-fixed and paraffin-embedded murine tissues were deparaffinized and incubated for 30 min in 3% hydrogen peroxide/methanol to quench endogenous peroxidases. Sections were rinsed in PBS and immunostained with anti-(murine) SPL antisera at 1:200 dilution in 0.5% PBS/ova albumin at 37°C for 1 h after antigen retrieval with citrate buffer (pH 6.0) in a small autoclave set for 125°C for 2 min; slides were cooled for 1 h at RT before adding secondary antibody. Secondary antibody was biotinylated anti-rabbit (Vector laboratories) diluted 1:1,000 in 0.5% PBS/ova albumin and incubated for 30 min at RT. Sections were incubated with Elite ABC kit (Vector Laboratories) for 30 min and rinsed in PBS, and detection was performed with DAB (Vector Laboratories) for 2 min and counterstained in hematoxylin. Tissues from a SPL knockout mouse were used as a negative control for IHC studies.
Flow cytometry
Blood cells and splenocytes were pooled and labeled with fluorescent antibodies including the B cell marker B220-APC, T cell marker CD3-FITC, polymorphonuclear cell marker Gr-1 PerCP Cy5.5, and monocyte marker Cd11b-PE, all obtained from eBiosciences, Inc. (San Diego, CA). Cell separation was performed on a Becton Dickinson FACSAria I cell sorter using FACSDiva software. Granulocytes were identified as CD11b+ and Gr-1+, whereas monocytes were identified as Cd11b+ and Gr1−. Cells were homogenized and used for immunoblotting.
Immunoblotting
Immunoblotting was performed using a rabbit polyclonal antibody generated against the C-terminal peptide of murine SPL of the sequence: C-VTQGNQMNGSPKPR. Murine SPL antibody was used at 1:1,000 dilution. Actin antibody (Sigma) was used as a loading control at 1:20,000 dilution. Immunoblotting was conducted essentially as described previously (47).
SPL enzyme activity assays
SPL enzyme activity was performed using tritium radiolabeled dihydrosphingosine-1-phosphate substrate as described previously (57).
Relative levels of tissue SPL expression by IHC and frozen section β-Gal staining
To provide a rough estimate of total organ SPL expression levels that would allow for comparison with SPL activity levels in different organs, we used a scoring system for β-Gal or SPL IHC. This scoring system reflected the staining intensity level and the distribution of stain (percentage of cross-sectional area that expresses SPL at any given level) within an organ. A full cross-section of the organ was assessed, thereby providing information incorporating the signal from many substructures. This represents a modified combined intensity and distribution score (58). In other words, the score for an organ reflects total expression levels that can be affected by high, intense expression in specific substructures as well as low but detectible expression (as compared with the appropriate negative control tissue) in large areas of tissue. The tissue with the most robust cell staining, small intestinal villous tips, was uniformly and strongly positive and was thus assigned a value of 1.0 (i.e., 100% value). All other tissues were scored relative to this reference tissue. For example, the thymus in total would give a low score due to the low to undetectable staining in immature lymphocytes. However, because the thymic epithelium was uniformly stained with moderate to high intensity, the score for thymic tissue was very high (0.95). In contrast, a minority of heart muscle cells exhibited consistently weak staining intensity, and other heart muscle cells showed no stain, resulting in a score of 0.05.
Embryo isolation
C57BL/6 mice heterozygous for the Sgpl1 null allele were mated to generate litters containing some SPL knockout embryos and some embryos WT for SPL. Embryos were removed from the uterus and yolk sac of pregnant mice at various days of gestation as described (59). Genotyping was performed to identify knockout embryos that were used as negative controls for IHC staining. Representative parasagittal sections of E17 embryos were stained for SPL IHC as described below.
RESULTS
Generation of β-Gal SPL reporter mice
Global disruption of murine SPL expression results in 100% lethality in the weaning period (31, 39). Therefore, to investigate tissue-specific SPL functions in the adult mouse, we used a combination of recombineering and Cre-lox technologies to generate an SPL conditional knockout mouse model. To facilitate the analysis of SPL expression in different murine tissues, the targeting vector was designed to introduce a bacterial LacZ gene 5′ to exon 9, thereby creating a β-Gal reporter under control of the Sgpl1 gene regulatory region and resulting in SPL truncation (SPL reporter mice). Mice heterozygous for the SPL allele containing the reporter are born at expected mendelian frequency, exhibit no developmental or other phenotypes, and have normal reproductive function and lifespan, similar to mice heterozygous for a Sgpl1 null allele. These reporter mice were used to characterize SPL expression as described below. The targeting cassette that harbored the reporter gene was deleted to generate a Sgpl1 allele in which exons 9–11 containing the enzyme cofactor-binding site are “floxed” for the establishment of tissue-specific SPL knockout mice lines described elsewhere (A. Pandurangan, et al., unpublished observations).
Whole mount survey of β-Gal stained adult SPL reporter mouse organs
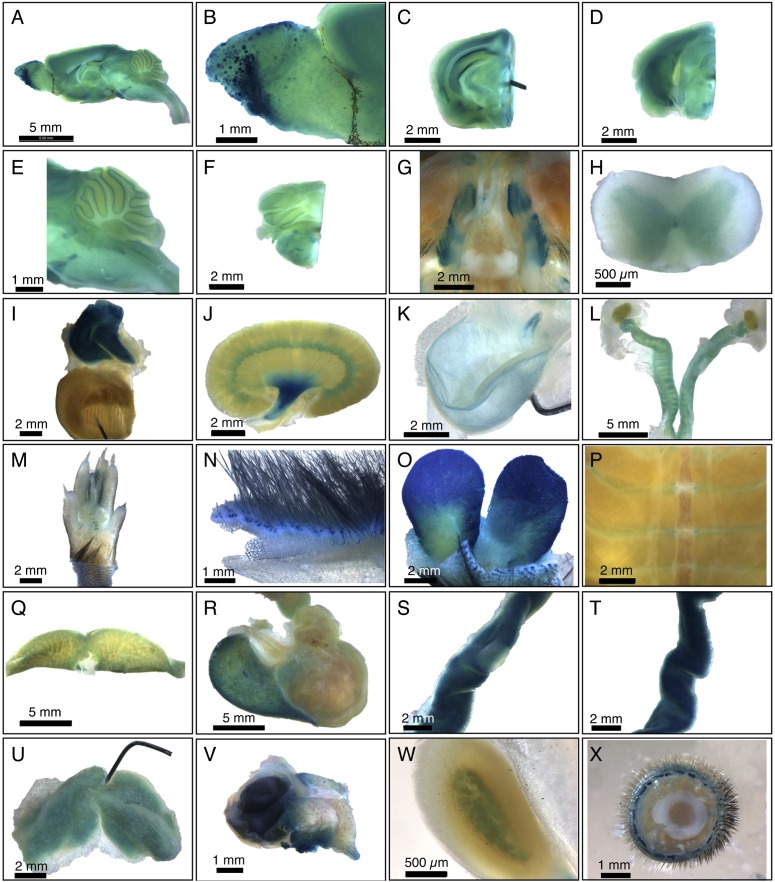
To undertake a comprehensive survey of SPL expression in the adult mouse, we performed whole mount analysis of β-Gal expression in organs dissected from male and female SPL reporter mice. SPL exhibits a broad range of expression levels in adult tissues. Little or no expression is detected in the whole mount analysis of tongue, spleen, pancreas, quadriceps muscle, sciatic nerve, heart atrium and ventricle, lung, aorta, diaphragm, mammary gland, pituitary gland, lacrimal gland, mesenteric adipose tissue, penis, prostate, vesicular gland, trachea, esophagus, and duodenum (data not shown). Staining in some tissues such as the salivary gland appears to be nonspecific, and faint expression is found in the thyroid gland (data not shown). In contrast, significant expression is observed in brain, spinal cord, trigeminal nerve ganglion, thymus, kidney, bladder, skin, preputial gland, Harderian gland, ribcage, liver, stomach, jejunum, ileum, cecum, colon, brown adipose tissue, adrenal cortex, and male and female reproductive organs (Fig. 1A–X). In some tissues, including thymus, preputial gland, Harderian gland, jejunum, and ileum, β-Gal expression is strong and uniform. In other cases, staining reveals specific patterns of expression, such as in the cortex, medulla, and papilla of the kidney, discrete glomeruli of the olfactory bulb, specific regions of the hippocampus, midbrain, hindbrain and granular layers of cerebellum, spinal cord gray matter, costal cartilage of the ribcage, fundus of the stomach, medulla of the adrenal gland, hair follicles of the skin, and epithelium of the bladder.
Fig. 1.
Whole mount survey of β-Gal-stained adult SPL reporter mouse organs. A: Whole brain. B: Olfactory bulb, showing staining of specific olfactory ganglia. C: Forebrain. D: Midbrain. E: Cerebellum and brainstem. F: Hindbrain. G: Basicranium, showing positively stained trigeminal nerve ganglia and unstained pituitary. H: Spinal cord, showing gray matter staining. I: Thymus and heart. Thymus is intensely stained. The pin is within the ventricular wall of the bisected heart, and the view shows the interior of the ventricle. J: Kidney, showing strong staining in the renal pelvis. K: Bladder, showing epithelial staining. L: Female gonads, including ovaries, fallopian tubes, and uterus. M: Foot. N: Skin. O: Preputial gland. P: Ribcage. Q: Liver. R: Stomach. S: Jejunum, with a Peyer's Patch in the center and showing less staining than surrounding jejunal tissue. T: Ileum. U: Brown adipose tissue. V: Harderian gland and eye. :) Adrenal gland. X: Tail. Data are representative of results obtained from four animals (two male and two female).
Histological analysis of SPL reporter mouse tissues by β-Gal staining
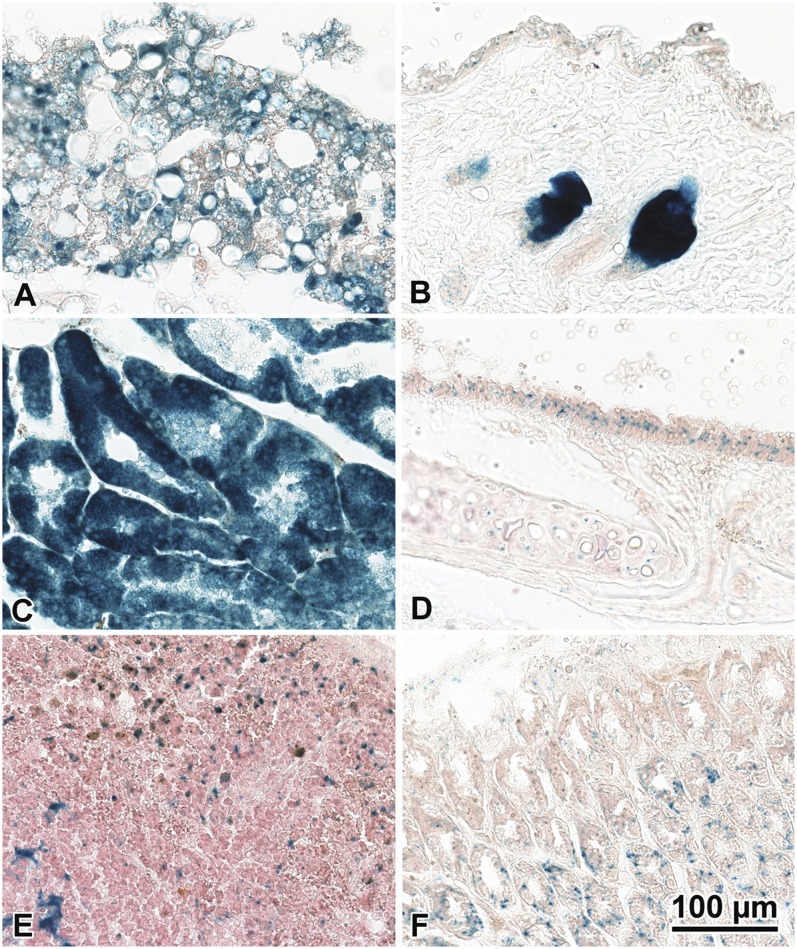
To gain more detailed analysis of some organs that stained strongly in whole mount survey, selected tissues were fixed and stained for β-Gal. Brown fat exhibits strong SPL expression (Fig. 2A). In the skin, the sebaceous glands are strongly positive (Fig. 2B), and weak staining is detected in the squamous epithelium. The Harderian gland of the eye stains strongly (Fig. 2C). The tracheal epithelium, spleen, and glandular stomach show some SPL expression (Fig. 2D–F).
Fig. 2.
β-Gal staining of SPL reporter mouse tissues. Adult mouse tissues were fixed and stained for β-Gal as described in Materials and Methods. A: Brown fat. B: Sebaceous glands and squamous epithelium of the skin. C: Harderian gland. D: Tracheal epithelium. E: Spleen. F: Stomach. Data are representative of results from three animals.
Survey of specific SPL adult tissues of interest by IHC
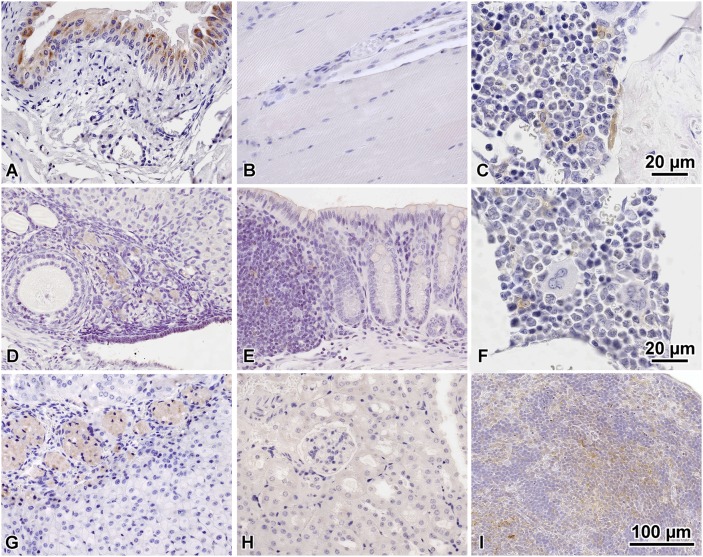
We next evaluated SPL expression by IHC in a range of tissues, including tissues exhibiting and lacking SPL expression as determined in the whole mount survey. Bladder transitional epithelium shows strong but patchy expression (Fig. 3A). Skeletal muscle is weakly positive, and there is a neurovascular bundle with similar weak positive staining in the nerve fibers (Fig. 3B). The bone marrow compartment shows specific immunoreactivity in the osteoclasts as well as rare cells with relatively abundant cytoplasm and immature nuclei located in the marrow itself, possibly representing marrow stromal cells (Fig. 3C, F). However, the majority of the hematopoietic cells are negative. The ovarian primordial oocytes adjacent to the primary follicle are positive for SPL, whereas the oocyte in the primary follicle is negative (Fig. 3D). Colon tissues show faint luminal membrane positivity in the epithelial cells as well as occasional cells in the gastrointestinal associated lymphoid tissue, possibly lymphoid stromal or dendritic cells (Fig. 3E). The adrenal x-zone cells show moderate expression levels (Fig. 3G). The kidney is weakly positive in some tubules (Fig. 3H). The spleen shows positive cells throughout the red pulp predominantly, with elongated cytoplasm likely representing splenic stromal supporting cells (Fig. 3I).
Fig. 3.
SPL IHC of WT adult mouse tissues. A: Bladder. B: Skeletal muscle. C: Bone marrow. D: Ovary. E: Colon. F: Bone marrow. G: Adrenal gland. H: Kidney. I: Spleen. Data are representative of results from three animals.
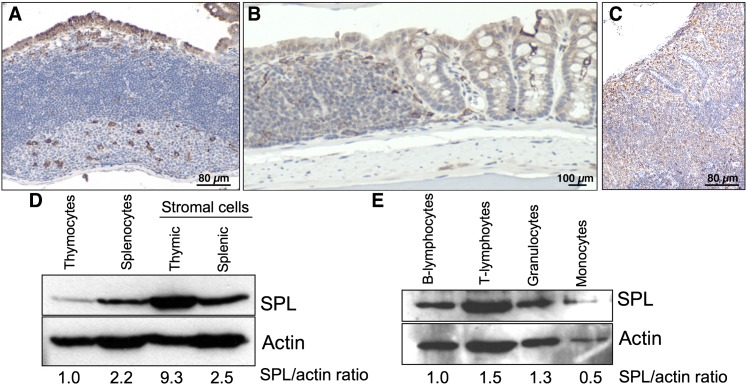
SPL expression in gut-associated lymphoid tissues, lymph nodes, and blood cells
Mice homozygous for the Sgpl1 null allele exhibit lymphopenia, retention of mature T cells in the thymus, and neutrophil trafficking defects. These findings suggest that SPL has multiple roles in innate and adaptive immunity. To provide a more in-depth analysis of SPL expression in immune cells and tissues, we evaluated SPL expression by IHC in gut-associated lymphoid tissues including Peyer's Patches of the jejunum (Fig. 4A) and colorectal lymphoid aggregates of the colon (Fig. 4B), as well as in peripheral lymph nodes (Fig. 4C). In addition, we used immunoblotting to compare the levels of SPL expression in peripheral blood leukocytes and the stromal and nonstromal fractions of thymus and spleen (Fig. 4D, E). SPL is expressed in each of the adult peripheral lymphoid organs (Fig. 4A–C). In the spleen, SPL is readily detected and expressed in splenocyte and stromal cell fractions with relatively equal intensities (Fig. 4D). In contrast, the stromal cells of the thymus exhibit a high level of SPL expression, whereas expression in the thymocyte fraction is barely detectible (Fig. 4D). These findings are consistent with our results from histological analysis of the thymus using IHC and β-Gal methodologies. SPL is also expressed in circulating blood cells, including B and T lymphocytes and granulocytes, with lower but detectible expression in monocytes (Fig. 4E).
Fig. 4.
SPL expression in gut-associated lymphoid tissues, lymph nodes, and blood cells. A: Peyer's Patch of the jejunum. B: Colorectal lymphoid aggregate. C: Lymph node. Data from A–C are representative of results from three animals. D: Thymocytes and splenocytes were isolated by mechanical dissociation of thymus and spleen, respectively. Remaining thymic and splenic stromal cells were washed twice with PBS. Cell homogenate was subjected to immunoblotting and probed with an SPL antibody and an actin antibody. Shown is a representative immunoblot with the relative expression of SPL in each cell population given below, as determined by ImageJ quantification of autoradiogram signal and shown as SPL signal normalized to actin signal. E: Leukocytes from spleen and whole blood were isolated, labeled with markers specific for each cell type indicated, and separated using a cell sorter as described in Materials and Methods. Cell homogenates from each fraction were subjected to immunoblotting and probed with SPL and actin antibodies. Shown is a representative immunoblot of SPL expression in T lymphocytes, B lymphocytes, granulocytes, and monocytes. The relative SPL expression of each cell population is given below, as determined by ImageJ quantification of autoradiogram signal and shown as SPL signal normalized to actin signal.
Survey of SPL IHC-positive brain tissues
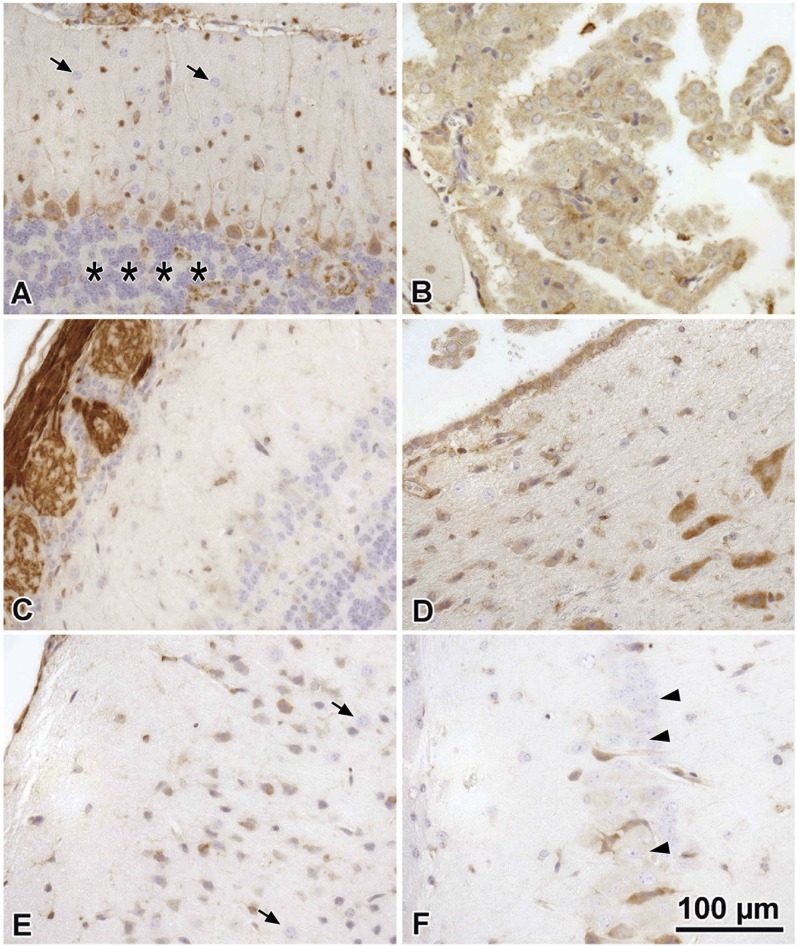
By histochemical analysis, the expression of SPL within each anatomical section of the brain appears highly specific, consistent with the distinct expression patterns observed in each brain compartment in the whole-mount β-Gal survey. The Purkinje layer neurons of the cerebellum are moderate in intensity but are uniformly immunoreactive (Fig. 5A). Cells lining the choroid plexus are positive (Fig. 5B). Olfactory neurons at the periphery of the olfactory bulb are one of the strongest SPL expressing tissues (Fig. 5C). The expression in the olfactory epithelium is found in the sensory neurons. SPL is expressed in the cell body proper and also in the axonal processes to the olfactory bulb. Brain stem neurons also demonstrate relatively strong SPL expression (Fig. 5D), comparable to the cerebellum and stronger than the cerebral cortical neurons, which are weakly to moderately positive. Meninges, particularly the arachnoid lining cells, are positive (Fig. 5E). Some of the thalamic nuclei neurons are positive, whereas others are negative for immunoreactivity (Fig. 5F). Glial astrocytes (Fig. 5F, most cells), oligodendroglia (5A top, and 5F smallest nuclei), granular cell layers of the cerebellum (Fig. 5A, bottom), and the majority of the brain monocytes/microglia are negative for immunoreactivity (not shown). Cell types in the brain were identified by morphologic features on standard stains including the hematoxylin counter staining in the IHC studies shown.
Fig. 5.
Survey of SPL IHC-positive brain tissues. A: Cerebellum showing positive Purkinje cells and processes and arachnoid meninges cells (top). Granular cell layers are nonimmunoreactive (asterisks), and cerebellar astrocytes are also negative (arrows). B: Choroid plexus lining cells are positive. C: Olfactory neurons and their processes (left) at the edge of the olfactory bulb are one of the strongest staining tissues. D: Brain stem neurons are relatively strong, comparable to the cerebellum and stronger than neurons of the cortex and thalamus. E: Meninges (left) and cerebral cortical neurons are moderately positive, whereas astrocytes are negative (arrows). F: Some individual thalamic nuclei cells are positive (arrowheads), whereas others are negative for immunoreactivity.
Validation and comparison of SPL expression detected by β-Gal reporter and IHC
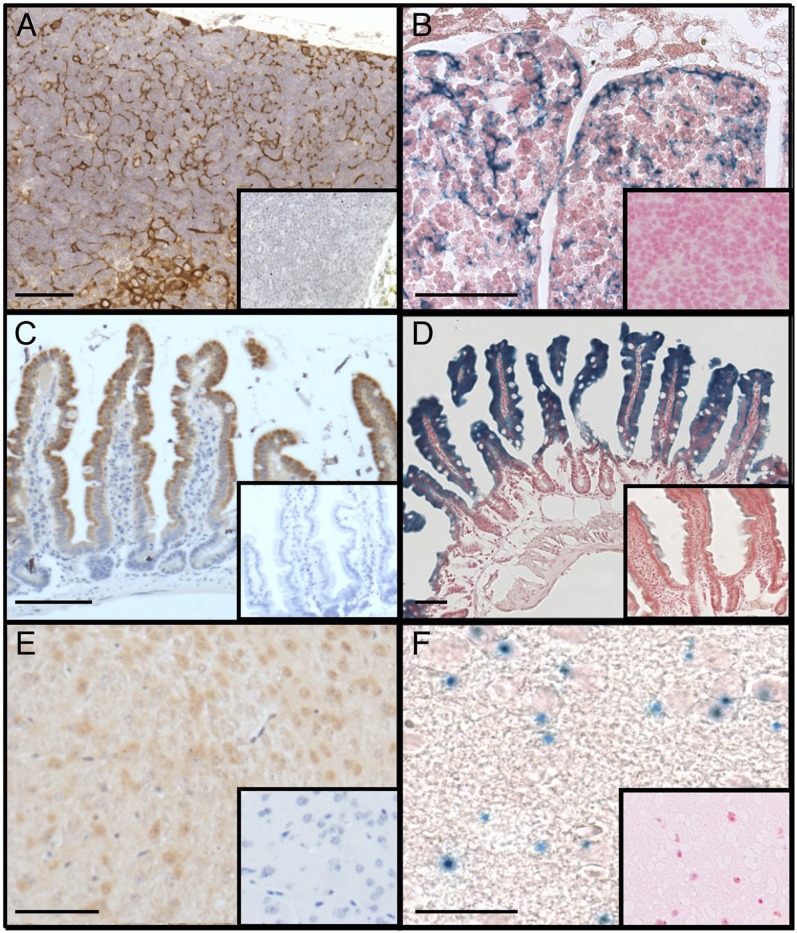
To establish if the SPL reporter is a reliable indicator of SPL expression, we compared histological results of β-Gal staining of (SPL reporter mouse) tissues known to express high levels of SPL with IHC staining of the corresponding tissues from a WT control mouse. For SPL IHC, we used a murine SPL antibody generated against a peptide corresponding to the C terminus of murine SPL. This antibody demonstrates a high degree of specificity, as shown by the absence of staining in tissues of mice homozygous for the Sgpl1 null allele. SPL knockout mouse tissues are the appropriate control for IHC studies, whereas tissues from a WT mouse lacking the SPL reporter insertion are the appropriate controls for β-Gal staining. Thymic epithelium of a WT mouse stains strongly using the SPL IHC detection system (Fig. 6A). In contrast, no staining is observed in the thymus of a SPL-null mouse (Fig. 6A, inset). SPL protein expression in the thymus determined by β-Gal staining shows corresponding patterns, with expression restricted to thymic epithelial cells of the stroma and little or no staining in thymocytes (Fig. 6B) and no staining in WT mouse tissues (Fig. 6B, inset). IHC of WT mouse jejunum shows strong staining in the differentiated enterocytes at the tip of the villi with less staining toward the base of the crypts (Fig. 6C), with no staining in SPL null mouse intestines (Fig. 6C, inset). Similarly, Fig. 6D shows robust β-Gal staining in the SPL reporter mouse jejunum, whereas no staining is detected in the WT mouse jejunum (Fig. 6D, inset). Brain cortical neurons exhibit much lower SPL expression levels. However, positive cells are similarly identified by both SPL detection systems but not in the corresponding negative control tissues (Fig. 6E, F, and corresponding insets). Overall, these results establish the specificity and sensitivity of IHC and β-Gal reporter systems to detect SPL expression in tissues expressing relatively high and low levels of SPL.
Fig. 6.
Validation of SPL detection systems and comparative IHC and β-Gal staining in adult mouse tissues. A: SPL IHC of thymus from a WT mouse. Inset shows lack of staining in tissues from a SPL knockout mouse. B: SPL expression by β-Gal staining of thymus from a SPL reporter mouse. Inset shows lack of staining in tissues from a WT mouse lacking the SPL reporter insertion. C: SPL expression by IHC of small intestinal villi from a WT mouse. Inset shows lack of staining from a SPL knockout mouse. D: β-Gal staining of small intestinal villi from a SPL reporter mouse. Inset shows lack of staining in tissues from a WT mouse. E: SPL expression by IHC of brain cortical neurons from a WT mouse. Inset shows lack of staining from a SPL knockout mouse. F: SPL expression by β-Gal staining of brain cortical neurons from a SPL reporter mouse. Inset shows lack of staining in tissues from a WT mouse. Bar = 100 µm.
Comparison of SPL expression among different tissues
To provide a comparison of SPL activity among different tissues of the adult mouse, we performed SPL enzyme activity assays in a range of organs from three individual mice (Fig. 7A). SPL activity was highest in the small intestine (jejunum), with lowest levels in heart and brain and intermediate levels in colon and thymus. We observed consistent levels of SPL enzyme activity within the same tissues of different individual mice. To compare SPL activity and SPL expression levels as determined by our different histological detection systems, we used a method that allows a rough estimate of total organ expression levels using each staining technique as described in Materials and Methods and as published previously (58). This method reflects a combined intensity and distribution score. The score for each organ was normalized to the most intense and consistent area of expression, the intestinal villi (Fig. 7B, C), which was arbitrarily set at 1. Strong IHC staining was observed in the intestines of adult SPL reporter mice (100%). Based on this reference tissue, uniform staining was also observed in the thymus (95%). In the stomach, 50% positive cells were observed. Much lower levels of SPL expression were found in the liver (30%), colon (20%), brain (15%), lung (10%), kidney (10%), and heart (5%). Relative expression of SPL, as indicated by β-Gal staining of tissues harvested from adult SPL reporter mice, is summarized in Fig. 7C. Based on strong positive staining in the intestines (considered 100% reference tissue), SPL was also robustly expressed in the thymus (95%) and spleen (90%). Staining in the stomach was 40%. Much lower levels of SPL expression were found in the liver (20%), brain (15%), colon (15%) lung (10%), kidney (10%), and heart (5%). There was good correlation between the enzyme activity, β-Gal, and IHC results in all of the above tissues. In contrast, IHC staining in the spleen was low (25%) compared with β-Gal activity (90%) and restricted to scattered splenic dendritic cells. This suggests a short lived/rapid turnover of the SPL protein compared with the stability of β-Gal or a source of falsely positive β-Gal activity in the spleen, although this was not seen in controls supporting the former conclusion.
Fig. 7.
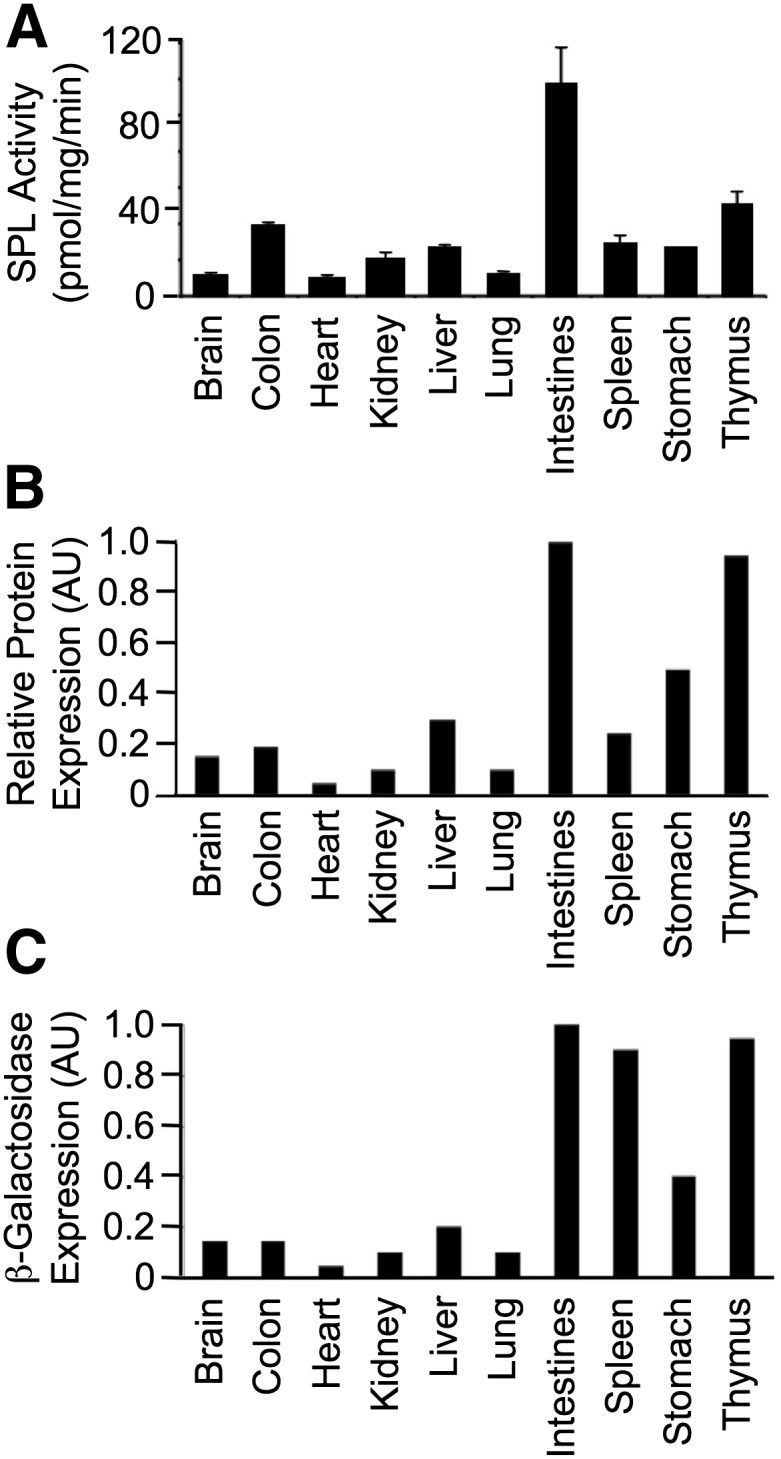
Relative SPL expression and activity in different adult mouse tissues. A: SPL enzyme activity levels. B: Relative SPL protein expression determined by quantitative analysis of IHC using polyclonal antimurine SPL antibody. C: Relative β-Gal staining intensity determined by quantitative analysis of β-Gal staining. AU, arbitrary units.
Expression of SPL during murine development
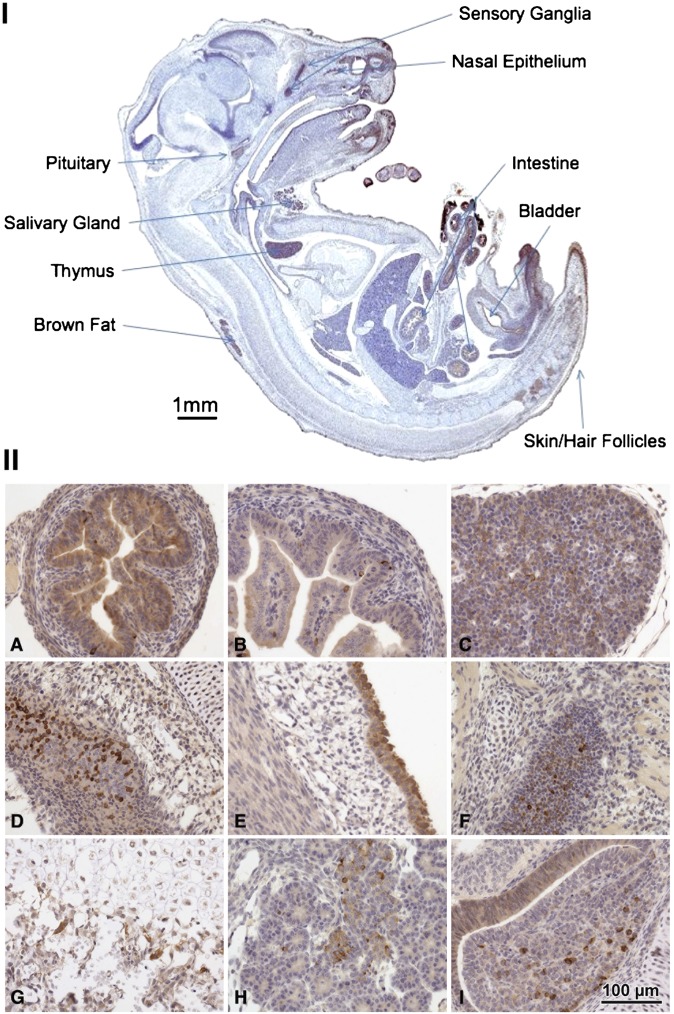
The expression of SPL was investigated in day 17 murine embryos (Fig. 8I). As in the adult, the intestinal epithelium and thymus exhibit the strongest expression of all the tissues. Positive cells are also noted in the developing sensory ganglia, olfactory lobe, nasal epithelium, salivary gland, bladder epithelium, pituitary gland, brown fat, skin, and hair.
Fig. 8.
Late-stage embryo tissues. (I) Midsection E18 with highest areas of expression indicated by labels. (II) Histological sections showing the strongest and most specific expression. A: Mid-small intestine, jejunum. B: Proximal small intestine, duodenum. C: Thymus. D: Upper respiratory mucosa and olfactory epithelium. E: Bladder transitional epithelium. F: Trigeminal ganglion. G: Bone epiphysis. H: Pancreas. I: Pituitary gland. Data are representative of results from three late-stage embryos.
Histological analysis of the tissues exhibiting the strongest and most specific staining are shown in Fig. 8IIA–I. The mid-small intestine (jejunum) shows diffuse strong epithelial staining with additional SPL-positive cells scattered in the basal layers. The proximal small intestine and duodenum show weaker luminal epithelial staining but still have scattered strong basal cells. The embryonic thymus is strongly positive, with a high density of the positive signal in thymic epithelial cells and a low signal in the lymphocytes. The upper respiratory mucosa and olfactory epithelium express high levels of SPL. The bladder transitional epithelium is strongly positive with much weaker expression in the smooth muscle. The trigeminal ganglion cells show scattered cells with strong expression. The developing bone at the epiphysis shows extensive staining in the osteoblasts. The developing exocrine pancreas is positive predominantly in ductal cells and more weakly in acinar cells. The pituitary gland shows scattered strongly positive cells in the adenohypophysis.
DISCUSSION
SPL is required for mammalian developmental and physiological functions, as shown by the congenital abnormalities, immunological and metabolic dysregulation, runting, and short lifespan observed in SPL null mice (39). To gain more insight into possible functions of SPL in the developing and adult mouse, we examined the spatial expression pattern of murine SPL using a combination of β-Gal reporter whole mount and histological stains, immunohistochemisty, and enzyme activity assays. A high degree of correlation was found between the three detection systems, demonstrating that the results are robust and validating the SPL reporter as a reliable system for SPL expression. A limited number of tissues, including the pituitary gland and spleen, gave discrepant results between different detection methods. This may be explained by poor penetration of detection reagents in the whole mount assay or the expression of SPL in specific cell types deep within the tissues that are not evident in whole mount assay. Some differences were also observed in intensity of SPL expression as determined by IHC versus SPL reporter. Although this could reflect technical conditions, such as the efficiency of antigen retrieval in different tissues with IHC detection, positional effects on the lacZ reporter, or endogenous β-Gal activity, the evaluation of multiple animals (both control and reporter/knockout) suggests that the differences are not based on artifact. Instead, it seems likely that some tissues have higher levels of transcription as detected by the lacZ reporter, even without higher levels of protein as detected by IHC. This suggests that there is post-transcriptional regulation or more rapid protein degradation in some tissues. Overall, a combination of detection systems, biochemical assays, and appropriate negative controls provides the most reliable information regarding SPL expression and function in individual tissues of interest.
Our results in small intestines, which express the highest levels of SPL among all the organs evaluated, are consistent with previous reports (47, 48). SPL expression in intestinal and colonic epithelia likely represents its role in the metabolism of dietary sphingolipids (60). Down-regulation of SPL in neoplastic intestinal tissues, combined with the established role of SPL in regulation of apoptosis, suggests that SPL may also facilitate the turnover of damaged intestinal epithelial cells.
Similarly, SPL is highly expressed in the thymus, corroborating previous reports (61, 62). SPL expression is restricted to the epithelial cells of the thymic stroma. It has been suggested that SPL expression in this location serves to maintain low thymic S1P levels, thereby generating the S1P gradient that facilitates mature lymphocyte egress, a concept that has not been established experimentally (42). SPL expression was not detected in thymic lymphocytes using β-Gal, IHC, or immunoblotting methods; nor was it detected by IHC in blood cells of the bone marrow. These findings, in combination with the notable SPL expression in peripheral lymph nodes, gut-associated lymphoid tissues, and circulating leukocytes, suggest that SPL expression may be induced in lymphocytes and other blood cells as they reach maturity. More precise studies of SPL expression during the stages of lymphocyte maturation and in other blood cell populations are required to confirm this possibility.
A lower-than-expected frequency of SPL homozygous null mice is observed in crosses of mice heterozygous for the null allele, and congenital anomalies, including runting, vascular and kidney defects, and thoracic malformations of the sternum, ribs and vertebrae, were observed (39). These findings are consistent with our observation that SPL is strongly expressed in specific regions of the adult and developing kidney and ribcage cartilage. The function of SPL in these locations is not known but may relate to the role of S1P in vascular maturation and/or cell migration, which has been shown to be defective in SPL null mouse embryonic fibroblasts. The pattern of SPL expression observed in the day 17 embryo indicates that the adult SPL expression pattern is established well before birth. Analysis of SPL expression in early-stage embryos will provide more insight regarding the contribution of SPL function to the development and/or prenatal functions of specific organs.
Some of the SPL expression patterns we observed in developing and adult mice have not been previously reported or examined histologically. For example, SPL appears to be highly expressed in sebaceous glands of the skin, with lower but detectible expression in skin epithelial cells. This finding is interesting in light of previous reports that Sgpl1 expression is altered in atopic skin disease in canines and humans (63, 64). Additionally, the fatty acid dehydrogenase whose deficiency is responsible for the skin disorder Sjogren-Larsson syndrome was recently shown to catabolize the SPL product hexadecenal (65). Finally, a recent study implicated SPL and its impact on S1P metabolism and calcium homeostasis as potentially playing a role in Darier's disease, which is caused by a defect in keratinocyte adhesion and differentiation (64). Our finding of SPL expression in discrete glomeruli of the olfactory bulb and embryonic olfactory mucosa is consistent with reports demonstrating Sgpl1 expression and activity in this location by other methods (66, 67). Combined with the finding of high SPL expression in the preputial gland, which is responsible for synthesis of pheromones, these findings suggest that SPL may play a role in olfactory organ function and the biosynthesis, metabolism, and/or detection of pheromones. We recently showed that one of the products of the SPL reaction, hexadecenal, has biological activity and activates JNK signaling (52). Interestingly, some long-chain aldehydes function as pheromones, which suggests the possibility that SPL may be required for the production of hexadecenal in olfactory organs (68–70). Our finding of SPL expression in the Purkinje cell layer of the cerebellum is consistent with a recent study by Hagen and colleagues (71). SPL expression in other specific neurons of the forebrain, midbrain, hindbrain, cerebellum, spinal cord, and trigeminal ganglion have not been previously reported and suggest that SPL may have important functions in these locations. This notion is supported by the observation that SPL-null mouse neurons undergo degeneration and that neurons that robustly express SPL are the first to degenerate in SPL-null mice (71). SPL expression is critical for preventing S1P-induced apoptosis in hippocampal neurons, and sphingosine kinases are required for normal brain development (17, 71, 72). These observations suggest the need for tight regulation of S1P levels in the developing and adult brain. SPL expression in the dorsal root ganglia of the spinal cord, which contains the cell bodies of afferent spinal nerves, is interesting in light of the finding that S1P and other lysophospholipid mediators are important in nocioception (73, 74). The observation that SPL is highly expressed in brown adipose tissue is interesting in light of the previous finding that SPL-null mice are lean, despite exhibiting high tissue and circulating lipid levels (31). SPL expression in the liver is consistent with the finding of metabolic and immune dysregulation in liver tissues of SPL-null mice (31). The finding of SPL expression in the developing and adult bladder epithelium and bronchial epithelium has not been previously reported. SPL expression in these locations and in the epithelium of the gut could indicate tissue-specific functions involved in maintenance of barrier integrity, immune defenses, and/or promotion of injured cell turnover.
The levels of S1P among different rodent tissues have been quantitated previously by several groups using different biochemical methods. In 1997, Yatomi (75) showed that S1P levels in rat tissues varied from lowest in liver, heart, and skeletal muscle to highest in intestines, with intermediate levels in brain, kidney, and spleen. S1P levels were not measured in the thymus (which we measured) but were high in testis (which we did not measure). In a study by Jiang and Han (76), S1P was determined in brain compartments and was found to be highest in spinal cord and brainstem, with lowest levels in cortex and intermediate levels in cerebellum. These findings show that SPL expression roughly correlates positively with S1P levels. This could be interpreted in two ways. SPL could have little bearing on S1P levels because its high expression does not result in low S1P levels. Alternatively, and most likely, high SPL expression is required in tissues subjected to high S1P biosynthesis or uptake. Mice with reduced global SPL activity exhibit high levels of S1P in most tissues in comparison to control mice, as shown by others and ourselves (31, 40, 42, 44, 46). Even in tissues such as heart and skeletal muscle, wherein baseline SPL levels are very low to undetectable, SPL inhibition has significant effects on S1P levels. More detailed analysis of the impact of SPL expression in different tissues will require the conditional disruption of SPL in specific organs and tissues. Gross changes in tissue S1P levels may not accurately reflect the importance of highly localized gradients that influence the viability and migration of discrete cell populations. In addition, S1P levels may be controlled by S1P phosphatases, such as has been demonstrated in the thymus (29).
SPL expression was not detectible in some tissues, notably the heart and skeletal muscles. However, we have shown that SPL is induced in the murine heart and skeletal striated muscle in response to ischemia and chemical injury, respectively (44, 46). SPL expression is regulated by transcriptional and post-transcriptional mechanisms during development and in response to radiation, ischemia and other insults. Thus, it is important to recognize that the baseline SPL expression patterns we observe likely underestimate the functions of SPL, especially those that may come into play under stressful conditions in which apoptosis is exacerbated and tissues are subject to injury or degeneration.
Acknowledgments
The authors thank the LacZ team at the UC Davis Mouse Biology Program for excellent technical assistance and expertise and John Ngai and Russell Fletcher for helpful discussions.
Footnotes
Abbreviations:
- β-Gal
- β-galactosidase
- IHC
- immunohistochemistry
- RT
- room temperature
- S1P
- sphingosine-1-phosphate
- S1PR
- sphingosine-1-phosphate receptor
- SPL
- sphingosine-1-phosphate lyase
- WT
- wild-type
This work was supported by National Institutes of Health Grants CA77528, GM66954, CA129438 (J.D.S.); K26 RR024037 and U01 CA14582 (A.D.B.); U42RR033193 and U54HG006364 (D.W.); and P01 HL090553, HL76839, HL86683, and GM66152 and by March of Dimes grant 6-FY2007-1012 and an Ellison Medical Foundation Senior Scholar Award (S.G.Y.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Hla T. 2003. Signaling and biological actions of sphingosine 1-phosphate. Pharmacol. Res. 47: 401–407. [DOI] [PubMed] [Google Scholar]
- 2.Maceyka M., Harikumar K. B., Milstien S., Spiegel S. 2012. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 22: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiegel S., Milstien S. 2003. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4: 397–407. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Wada R., Yamashita T., Mi Y., Deng C. X., Hobson J. P., Rosenfeldt H. M., Nava V. E., Chae S. S., Lee M. J., et al. 2000. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Invest. 106: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kono M., Belyantseva I. A., Skoura A., Frolenkov G. I., Starost M. F., Dreier J. L., Lidington D., Bolz S. S., Friedman T. B., Hla T., et al. 2007. Deafness and stria vascularis defects in S1P2 receptor-null mice. J. Biol. Chem. 282: 10690–10696. [DOI] [PubMed] [Google Scholar]
- 6.Kupperman E., An S., Osborne N., Waldron S., Stainier D. Y. 2000. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 406: 192–195. [DOI] [PubMed] [Google Scholar]
- 7.Kawahara A., Nishi T., Hisano Y., Fukui H., Yamaguchi A., Mochizuki N. 2009. The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science. 323: 524–527. [DOI] [PubMed] [Google Scholar]
- 8.Wong R. C., Tellis I., Jamshidi P., Pera M., Pebay A. 2007. Anti-apoptotic effect of sphingosine-1-phosphate and platelet-derived growth factor in human embryonic stem cells. Stem Cells Dev. 16: 989–1001. [DOI] [PubMed] [Google Scholar]
- 9.Nincheri P., Luciani P., Squecco R., Donati C., Bernacchioni C., Borgognoni L., Luciani G., Benvenuti S., Francini F., Bruni P. 2009. Sphingosine 1-phosphate induces differentiation of adipose tissue-derived mesenchymal stem cells towards smooth muscle cells. Cell. Mol. Life Sci. 66: 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inniss K., Moore H. 2006. Mediation of apoptosis and proliferation of human embryonic stem cells by sphingosine-1-phosphate. Stem Cells Dev. 15: 789–796. [DOI] [PubMed] [Google Scholar]
- 11.Avery K., Avery S., Shepherd J., Heath P. R., Moore H. 2008. Sphingosine-1-phosphate mediates transcriptional regulation of key targets associated with survival, proliferation, and pluripotency in human embryonic stem cells. Stem Cells Dev. 17: 1195–1205. [DOI] [PubMed] [Google Scholar]
- 12.Ye X. 2008. Lysophospholipid signaling in the function and pathology of the reproductive system. Hum. Reprod. Update. 14: 519–536. [DOI] [PubMed] [Google Scholar]
- 13.Rodgers A., Mormeneo D., Long J. S., Delgado A., Pyne N. J., Pyne S. 2009. Sphingosine 1-phosphate regulation of extracellular signal-regulated kinase-1/2 in embryonic stem cells. Stem Cells Dev. 18: 1319–1330. [DOI] [PubMed] [Google Scholar]
- 14.Ratajczak M. Z., Lee H., Wysoczynski M., Wan W., Marlicz W., Laughlin M. J., Kucia M., Janowska-Wieczorek A., Ratajczak J. 2010. Novel insight into stem cell mobilization-plasma sphingosine-1-phosphate is a major chemoattractant that directs the egress of hematopoietic stem progenitor cells from the bone marrow and its level in peripheral blood increases during mobilization due to activation of complement cascade/membrane attack complex. Leukemia. 24: 976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pebay A., Wong R. C., Pitson S. M., Wolvetang E. J., Peh G. S., Filipczyk A., Koh K. L., Tellis I., Nguyen L. T., Pera M. F. 2005. Essential roles of sphingosine-1-phosphate and platelet-derived growth factor in the maintenance of human embryonic stem cells. Stem Cells. 23: 1541–1548. [DOI] [PubMed] [Google Scholar]
- 16.Mizugishi K., Li C., Olivera A., Bielawski J., Bielawska A., Deng C. X., Proia R. L. 2007. Maternal disturbance in activated sphingolipid metabolism causes pregnancy loss in mice. J. Clin. Invest. 117: 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizugishi K., Yamashita T., Olivera A., Miller G., Spiegel S., Proia R. 2005. Essential role for sphingosine kinases in neural and vascular development. Mol. Cell. Biol. 25: 11113–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Hsu A., Lee J., Cramer D., Lee M. 2011. To stay or to leave: stem cells and progenitor cells navigating the S1P gradient. World J. Biol. Chem. 2: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera J., Proia R. L., Olivera A. 2008. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat. Rev. Immunol. 8: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H., Deng J., Kujawski M., Yang C., Liu Y., Herrmann A., Kortylewski M., Horne D., Somlo G., Forman S., et al. 2010. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat. Med. 16: 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordula T., et al. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 465: 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karliner J. S. 2002. Lysophospholipids and the cardiovascular system. Biochim. Biophys. Acta. 1582: 216–221. [DOI] [PubMed] [Google Scholar]
- 23.Morita Y., Perez G. I., Paris F., Miranda S. R., Ehleiter D., Haimovitz-Friedman A., Fuks Z., Xie Z., Reed J. C., Schuchman E. H., et al. 2000. Oocyte apoptosis is suppressed by disruption of the acid sphingomyelinase gene or by sphingosine-1-phosphate therapy. Nat. Med. 6: 1109–1114. [DOI] [PubMed] [Google Scholar]
- 24.Paris F., Perez G., Fuks Z., Haimovitz-Friedman A., Nguyen H., Bose M., Ilagan A., Hunt P., Morgan W., Tilly J., et al. 2002. Sphingosine-1-phosphate preserves fertility in irradiated female mice without propagating genomic damage in offspring. Nat. Med. 8: 901–902. [DOI] [PubMed] [Google Scholar]
- 25.Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., et al. 2009. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 325: 1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitson S. M. 2011. Regulation of sphingosine kinase and sphingolipid signaling. Trends Biochem. Sci. 36: 97–107. [DOI] [PubMed] [Google Scholar]
- 27.Pyne S., Lee S. C., Long J., Pyne N. J. 2009. Role of sphingosine kinases and lipid phosphate phosphatases in regulating spatial sphingosine 1-phosphate signalling in health and disease. Cell. Signal. 21: 14–21. [DOI] [PubMed] [Google Scholar]
- 28.Brindley D. N., Pilquil C. 2009. Lipid phosphate phosphatases and signaling. J. Lipid Res. 50(Suppl): S225–S230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bréart B., Ramos-Perez W. D., Mendoza A., Salous A. K., Gobert M., Huang Y., Adams R. H., Lafaille J. J., Escalante-Alcalde D., Morris A. J., et al. 2011. Lipid phosphate phosphatase 3 enables efficient thymic egress. J. Exp. Med. 208: 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serra M., Saba J. D. 2010. Sphingosine 1-phosphate lyase, a key regulator of sphingosine 1-phosphate signaling and function. Adv. Enzyme Regul. 50: 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bektas M., Allende M. L., Lee B. G., Chen W., Amar M. J., Remaley A. T., Saba J. D., Proia R. L. 2010. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J. Biol. Chem. 285: 10880–10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendel J., Heinecke K., Fyrst H., Saba J. D. 2003. Sphingosine phosphate lyase expression is essential for normal development in Caenorhabditis elegans. J. Biol. Chem. 278: 22341–22349. [DOI] [PubMed] [Google Scholar]
- 33.Gottlieb D., Heideman W., Zhou J., Oskouian B., Saba J. 1999. The DPL1 gene is involved in mediating the response to nutrient deprivation in Saccharomyces cerevisiae. Mol. Cell Biol. Res. Commun. 1: 66–71. [DOI] [PubMed] [Google Scholar]
- 34.Li G., Foote C., Alexander S., Alexander H. 2001. Sphingosine-1-phosphate lyase has a central role in the development of Dictyostelium discoideum. Development. 128: 3473–3483. [DOI] [PubMed] [Google Scholar]
- 35.Oskouian B., Saba J. D. 2004. Death and taxis: what non-mammalian models tell us about sphingosine-1-phosphate. Semin. Cell Dev. Biol. 15: 529–540. [DOI] [PubMed] [Google Scholar]
- 36.Herr D. R., Fyrst H., Phan V., Heinecke K., Georges R., Harris G. L., Saba J. D. 2003. Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development. 130: 2443–2453. [DOI] [PubMed] [Google Scholar]
- 37.Van Veldhoven P. P., Gijsbers S., Mannaerts G. P., Vermeesch J. R., Brys V. 2000. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q22. Biochim. Biophys. Acta. 1487: 128–134. [DOI] [PubMed] [Google Scholar]
- 38.Zhou J., Saba J. 1998. Identification of the first mammalian sphingosine phosphate lyase gene and its functional expression in yeast. Biochem. Biophys. Res. Commun. 242: 502–507. [DOI] [PubMed] [Google Scholar]
- 39.Schmahl J., Raymond C. S., Soriano P. 2007. PDGF signaling specificity is mediated through multiple immediate early genes. Nat. Genet. 39: 52–60. [DOI] [PubMed] [Google Scholar]
- 40.Vogel P., Donoviel M. S., Read R., Hansen G. M., Hazlewood J., Anderson S. J., Sun W., Swaffield J., Oravecz T. 2009. Incomplete inhibition of sphingosine 1-phosphate lyase modulates immune system function yet prevents early lethality and non-lymphoid lesions. PLoS ONE. 4: e4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagdanoff J. T., Donoviel M. S., Nouraldeen A., Tarver J., Fu Q., Carlsen M., Jessop T. C., Zhang H., Hazelwood J., Nguyen H., et al. 2009. Inhibition of sphingosine-1-phosphate lyase for the treatment of autoimmune disorders. J. Med. Chem. 52: 3941–3953. [DOI] [PubMed] [Google Scholar]
- 42.Schwab S. R., Pereira J., Matloubian M., Xu Y., Huang Y., Cyster J. 2005. Lymphocyte sequestration through S1P lyase inhibition an disruption of S1P gradients. Science. 309: 1735–1739. [DOI] [PubMed] [Google Scholar]
- 43.Bagdanoff J. T., Donoviel M. S., Nouraldeen A., Carlsen M., Jessop T. C., Tarver J., Aleem S., Dong L., Zhang H., Boteju L., et al. 2010. Inhibition of sphingosine 1-phosphate lyase for the treatment of rheumatoid arthritis: discovery of (E)-1-(4-((1R,2S,3R)-1,2,3,4-tetrahydroxybutyl)-1H-imidazol-2-yl)ethanone oxime (LX2931) and (1R,2S,3R)-1-(2-(isoxazol-3-yl)-1H-imidazol-4-yl)butane-1,2,3,4-tetraol (LX2932). J. Med. Chem. 53: 8650–8662. [DOI] [PubMed] [Google Scholar]
- 44.Bandhuvula P., Honbo N., Wang G. Y., Jin Z. Q., Fyrst H., Zhang M., Borowsky A. D., Dillard L., Karliner J. S., Saba J. D. 2011. S1P lyase: a novel therapeutic target for ischemia-reperfusion injury of the heart. Am. J. Physiol. Heart Circ. Physiol. 300: H1753–H1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar A., Oskouian B., Fyrst H., Zhang M., Paris F., Saba J. D. 2011. S1P lyase regulates DNA damage responses through a novel sphingolipid feedback mechanism. Cell Death Dis. 2: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loh K. C., Leong W., Carlson M., Oskouian B., Kumar A., Fyrst H., Zhang M., Proia R., Hoffman E., Saba J. 2012. Sphingosine-1-phosphate enhances satellite cell activation in dystrophic muscles through a S1PR2/STAT3 signaling pathway. PLoS ONE. 7: e37218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oskouian B., Sooriyakumaran P., Borowsky A., Crans A., Dillard-Telm L., Tam Y., Bandhuvula P., Saba J. 2006. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is downregulated in colon cancer. Proc. Natl. Acad. Sci. USA. 103: 17384–17389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohno M., Momoi M., Oo M., Paik J., Lee Y., Venkataraman K., Ai Y., Ristimaki A., Fyrst H., Sano H., et al. 2006. Intracellular role for sphingosine kinase 1 in intestinal adenoma cell proliferation. Mol. Cell. Biol. 26: 7211–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramaswamy S., Ross K. N., Lander E. S., Golub T. R. 2003. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33: 49–54. [DOI] [PubMed] [Google Scholar]
- 50.Colie S., Codogno P., Levade T., Andrieu-Abadie N. 2010. Regulation of cell death by sphingosine 1-phosphate lyase. Autophagy. 6: 426–427. [DOI] [PubMed] [Google Scholar]
- 51.Colie S., Van Veldhoven P. P., Kedjouar B., Bedia C., Albinet V., Sorli S. C., Garcia V., Djavaheri-Mergny M., Bauvy C., Codogno P., et al. 2009. Disruption of sphingosine 1-phosphate lyase confers resistance to chemotherapy and promotes oncogenesis through Bcl-2/Bcl-xL upregulation. Cancer Res. 69: 9346–9353. [DOI] [PubMed] [Google Scholar]
- 52.Kumar A., Byun H. S., Bittman R., Saba J. D. 2011. The sphingolipid degradation product trans-2-hexadecenal induces cytoskeletal reorganization and apoptosis in a JNK-dependent manner. Cell. Signal. 23: 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chipuk J. E., McStay G. P., Bharti A., Kuwana T., Clarke C. J., Siskind L. J., Obeid L. M., Green D. R. 2012. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 148: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams N., Gale N. 2006. High resolution gene expression analysis in mice using genetically inserted reporter genes. In Mammalian and avian transgenesis-new approaches. S. Pease and C. Lois, editors. Springer-Verlag, Berlin and Heidelberg, Germany. 131–172. [Google Scholar]
- 56.Ma W., Rogers K., Zbar B., Schmidt L. 2002. Effects of different fixatives on b-galactosidase activity. J. Histochem. Cytochem. 50: 1421–1424. [DOI] [PubMed] [Google Scholar]
- 57.Van Veldhoven P. P., Mannaerts G. P. 1991. Subcellular localization and membrane topology of sphingosine-1-phosphate lyase in rat liver. J. Biol. Chem. 266: 12502–12507. [PubMed] [Google Scholar]
- 58.Harvey J. M., Clark G. M., Osborne C. K., Allred D. C. 1999. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J. Clin. Oncol. 17: 1474–1481. [DOI] [PubMed] [Google Scholar]
- 59.Nagy A., Gertsenstein M., Vintersten K., Behringer R. R. 2003. Manipulating the mouse embryo: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 60.Duan R. D., Nilsson A. 2000. Sphingolipid hydrolyzing enzymes in the gastrointestinal tract. Methods Enzymol. 311: 276–286. [DOI] [PubMed] [Google Scholar]
- 61.Sensken S. C., Bode C., Nagarajan M., Peest U., Pabst O., Graler M. H. 2010. Redistribution of sphingosine 1-phosphate by sphingosine kinase 2 contributes to lymphopenia. J. Immunol. 184: 4133–4142. [DOI] [PubMed] [Google Scholar]
- 62.Weber C., Krueger A., Munk A., Bode C., Van Veldhoven P. P., Graler M. H. 2009. Discontinued postnatal thymocyte development in sphingosine 1-phosphate-lyase-deficient mice. J. Immunol. 183: 4292–4301. [DOI] [PubMed] [Google Scholar]
- 63.Seo E. Y., Park G. T., Lee K. M., Kim J. A., Lee J. H., Yang J. M. 2006. Identification of the target genes of atopic dermatitis by real-time PCR. J. Invest. Dermatol. 126: 1187–1189. [DOI] [PubMed] [Google Scholar]
- 64.Celli A., Mackenzie D. S., Zhai Y., Tu C. L., Bikle D. D., Holleran W. M., Uchida Y., Mauro T. M. 2012. SERCA2-controlled Ca(2)-dependent keratinocyte adhesion and differentiation is mediated via the sphingolipid pathway: a therapeutic target for Darier's disease. J. Invest. Dermatol. 132: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakahara K., Ohkuni A., Kitamura T., Abe K., Naganuma T., Ohno Y., Zoeller R. A., Kihara A. 2012. The sjogren-larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol. Cell. 46: 461–471. [DOI] [PubMed] [Google Scholar]
- 66.Genter M. B. 2006. Molecular biology of the nasal airways: how do we assess cellular and molecular responses in the nose? Toxicol. Pathol. 34: 274–280. [DOI] [PubMed] [Google Scholar]
- 67.Genter M. B., Van Veldhoven P. P., Jegga A. G., Sakthivel B., Kong S., Stanley K., Witte D. P., Ebert C. L., Aronow B. J. 2003. Microarray-based discovery of highly expressed olfactory mucosal genes: potential roles in the various functions of the olfactory system. Physiol. Genomics. 16: 67–81. [DOI] [PubMed] [Google Scholar]
- 68.Luo M. 2004. Got milk? A pheromonal message for newborn rabbits. Bioessays. 26: 6–9. [DOI] [PubMed] [Google Scholar]
- 69.Millar J. G. 2000. Polyene hydrocarbons and epoxides: a second major class of lepidopteran sex attractant pheromones. Annu. Rev. Entomol. 45: 575–604. [DOI] [PubMed] [Google Scholar]
- 70.Baum M. J., Kelliher K. R. 2009. Complementary roles of the main and accessory olfactory systems in mammalian mate recognition. Annu. Rev. Physiol. 71: 141–160. [DOI] [PubMed] [Google Scholar]
- 71.Hagen N., Hans M., Hartmann D., Swandulla D., van Echten-Deckert G. 2011. Sphingosine-1-phosphate links glycosphingolipid metabolism to neurodegeneration via a calpain-mediated mechanism. Cell Death Differ. 18: 1356–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hagen N., Van Veldhoven P. P., Proia R. L., Park H., Merrill A. H., Jr, Van Echten-Deckert G. 2009. Subcellular origin of sphingosine-1-phosphate is essential for its toxic effect in lyase deficient neurons. J. Biol. Chem. 284: 11346–11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue M., Rashid M. H., Fujita R., Contos J. J., Chun J., Ueda H. 2004. Initiation of neuropathic pain requires lysophosphatidic acid receptor signaling. Nat. Med. 10: 712–718. [DOI] [PubMed] [Google Scholar]
- 74.Doyle T., Chen Z., Obeid L. M., Salvemini D. 2011. Sphingosine-1-phosphate acting via the S1P(1) receptor is a downstream signaling pathway in ceramide-induced hyperalgesia. Neurosci. Lett. 499: 4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yatomi Y., Welch R. J., Igarashi Y. 1997. Distribution of sphingosine 1-phosphate, a bioactive sphingolipid, in rat tissues. FEBS Lett. 404: 173–174. [DOI] [PubMed] [Google Scholar]
- 76.Jiang X., Han X. 2006. Characterization and direct quantitation of sphingoid base-1-phosphates from lipid extracts: a shotgun lipidomics approach. J. Lipid Res. 47: 1865–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]