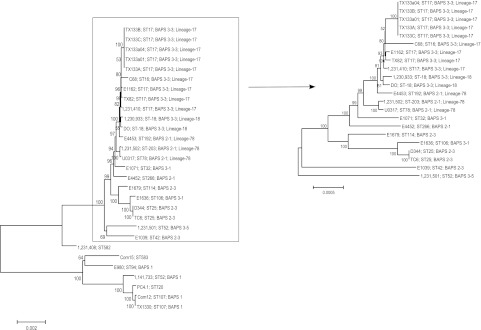
FIG 2 .
Minimum evolution tree based on the concatenated alignments of 299 orthologous proteins conserved in draft genome sequences of 29 E. faecium isolates. Bootstrap values are indicated and are based on 1,000 permutations. Strain codes are indicated as well as ST, BAPS group, and lineage based on their ST. Accession numbers: E. faecium 1231408, GenBank NZ_ACBB00000000; E. faecium 1231501, GenBank NZ_ACAY00000000; E. faecium TX0133A, GenBank NZ_AECH00000000; E. faecium TX0082, GenBank NZ_AEBU00000000; E. faecium C68, GenBank NZ_ACJQ00000000; E. faecium 1231410, GenBank NZ_ACBA00000000; E. faecium 1230933, GenBank NZ_ACAS00000000; E. faecium E4453, GenBank AEDZ00000000; E. faecium 1231502, GenBank NZ_ACAX00000000; E. faecium E4452, GenBank AEOU00000000; E. faecium D344, GenBank NZ_ACZZ00000000; E. faecium TC6, GenBank NZ_ACOB00000000; E. faecium Com15, GenBank NZ_ACBD00000000; E. faecium 1141733, GenBank NZ_ACAZ00000000; E. faecium PC4.1, GenBank NZ_ADMM00000000; E. faecium Com12, GenBank NZ_ACBC00000000; E. faecium TX1330, GenBank NZ_ACHL00000000; E. faecium E980, GenBank ABQA00000000; E. faecium E1039, GenBank ACOS00000000; E. faecium E1071, GenBank ABQI00000000; E. faecium E1162, GenBank ABQJ00000000; E. faecium E1636, GenBank ABRY00000000; E. faecium E1679, GenBank ABSC00000000; E. faecium U0317, GenBank ABSW00000000; E. faecium DO, GenBank NZ_ACIY00000000.1; E. faecium TX133a01, GenBank NZ_AECJ00000000.1; E. faecium TX133a04, GenBank NZ_AEBC00000000.1; E. faecium TX133B, GenBank NZ_AECI00000000.1; E. faecium TX133C, GenBank NZ_AEBG00000000.1. Strains 1231408, PC4.1, and Com15 lack a BAPS (sub)group designation because STs extracted from the genome sequences of these strains were not assigned yet at the time the BAPS analysis was performed. To improve resolution of the upper part of the tree, the top 22 non-BAPS 1 strains were separately clustered using the minimum evolution method.