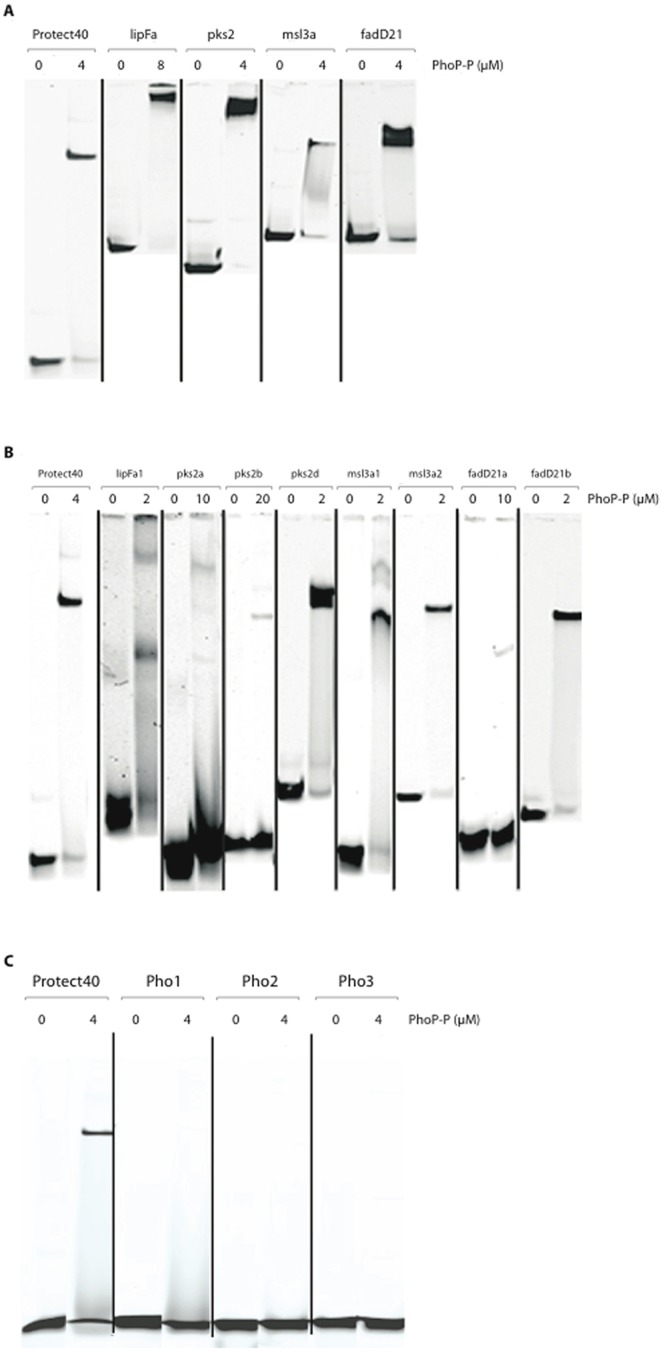
Figure 2. Electrophoretic mobility assays with the lipF, pks2, msl3 and fadD21 promoter fragments and PhoP-P.
2A. The large DNA fragments initially selected — lipFa (222 bp), pks2 (148 bp), msl3a (235 bp), and fadD21 (210 bp) — were incubated with PhoP-P in the presence of poly dI-dC at 10 µg/ml and run on a native polyacrylamide gel (8%), in 0.5× TBE buffer. Complete binding was assessed on the basis of the amount of shifted material for each fragment in the presence of 4 µM of PhoP-P (protein/DNA ratio: 100/1), except for lipFa, for which 8 µM PhoP-P was required (protein/DNA ratio: 200/1). 2B. Smaller DNA fragments — lipFa1 (68 bp), pks2a (59 bp), pks2b (40 bp), pks2d (104 bp), msl3a1 (45 bp), msl3a2 (83 pb), fadD21a (55 bp) and fadD21b (79 bp) — were designed and incubated with PhoP-P in the presence of poly dI-dC at 10 µg/ml. Protect40 (40 bp) was tested in all mobility shift assays, as a positive control [26]. 2C. Protect40 (40 bp) fragments with an altered sequence were used: Pho1 (DR1 altered sequence), Pho2 (DR2 altered sequence) and Pho3 (DR1 and DR2 altered sequences). Wild-type Protect40 (40 bp) was tested as a positive control [26].