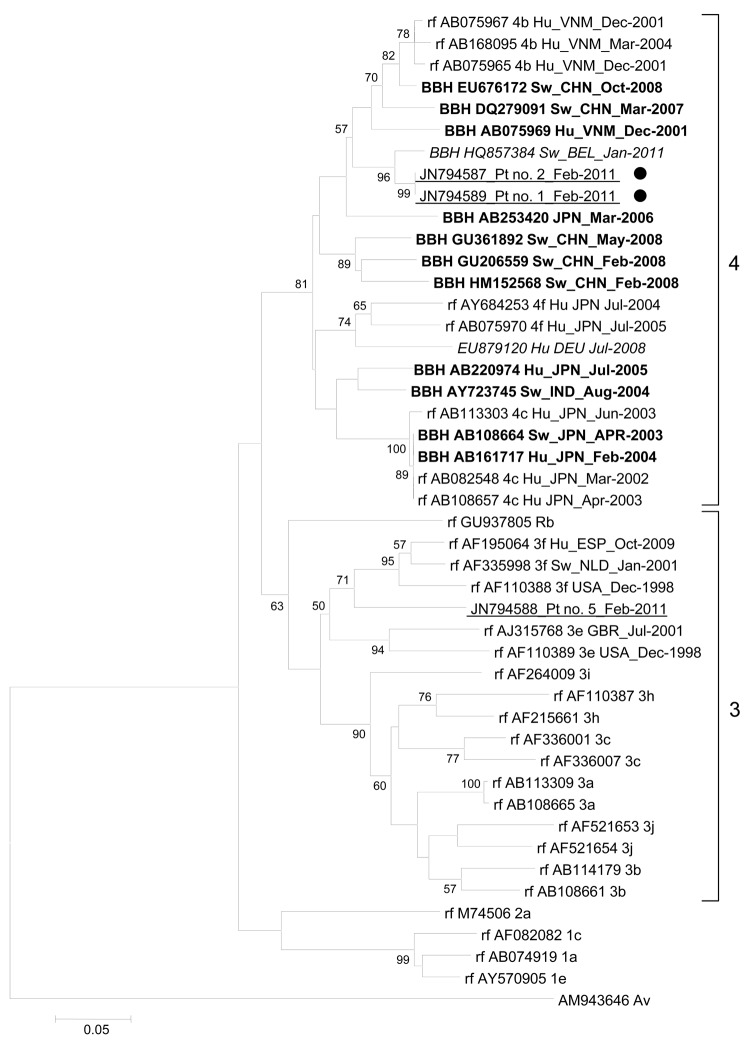
Figure.
Phylogenetic tree based on partial (186 nt) sequence of the 5′ end of open reading frame 1 of the hepatitis E virus (HEV) genome (nt 133–318; GenBank accession no. AB291961). Boldface indicates sequence with the highest BLAST scores (http://blast.ncbi.nlm.nih.gov/Blast.cgi); red (italics) indicates sequences obtained from a swine in Belgium with HEV genotype 4 (8) and a human in Germany with autochthonous HEV infection (9); blue (underlining) indicates sequences obtained from humans in Marseille, France, during January–March 2011; black dots indicate patients who ate uncooked pig liver sausage. Reference sequences (rf) with known genotypes and subtypes are indicated (6). Numbers on right indicate genotype. For sequences from this study, nucleotide alignments were performed by using ClustalX version 2.0 (www.clustal.org/download/current). The tree was constructed by using MEGA4 (www.megasoftware.net) and the neighbor-joining method as described (2). Branches were obtained from 1,000 resamplings of the data; those with bootstrap values >50% are labeled on the tree. The avian HEV sequence AM943646 was used as an outgroup. Scale bar indicates nucleotide substitutions per site. HEV sequences are labeled with GenBank accession number, host, country where isolated, and collection or submission date. Hu, human; VNM, Vietnam; BBH, best BLAST hit; Sw, swine; CHN, China; BEL, Belgium; Pt, patient; JPN, Japan; DEU, Germany; IND, India; Rb, rabbit; ESP, Spain; NLD, the Netherlands; USA, United States; GBR, United Kingdom; Av, avian.