Abstract
Parathyroid hormone (PTH) is an essential regulator of endochondral bone formation and an important anabolic agent for the reversal of bone loss. PTH mediates its functions in part by regulating binding of the bone-related activating transcription factor 4 (ATF4) to the osteoblast-specific gene, osteocalcin. The basic helix-loop-helix (bHLH) factors Twist1 and Twist2 also regulate osteocalcin transcription in part through the interaction of the C-terminal “box” domain in these factors and Runx2. In this study, we discovered a novel function of PTH: its ability to dramatically decrease Twist1 transcription. Since ATF4 is a major regulator of the PTH response in osteoblasts, we assessed the mutual regulation between these factors and determined that Twist proteins and ATF4 physically interact in a manner that affects ATF4 DNA binding function. We mapped the interaction domain of Twist proteins to the C-terminal “box” domain and of ATF4, to the N-terminus. Furthermore, we demonstrate that Twist1 overexpression in osteoblasts attenuates ATF4 binding to the osteocalcin promoter in response to PTH. This study thus identifies Twist proteins as novel inhibitory binding partners of ATF4 and explores the functional significance of this interaction.
Keywords: Twist1, ATF4, PTH, osteoblasts, osteocalcin
INTRODUCTION
Parathyroid hormone (PTH) is a major mediator of bone remodeling and an essential regulator of calcium homeostasis. Inappropriate production of PTH or the PTH-related peptide (PTHrP) accounts for the majority of causes of hypercalcemia. PPR (aka PTH-1R), the receptor for PTH and PTHrP, is expressed on osteoblasts and is a member of the superfamily of receptors coupled to guanyl nucleotide-binding regulatory G proteins. This receptor which signals through at least two second messenger systems, adenylate cyclase and phospholipase C, can be activated in a similar manner by the amino-terminal fragments of PTH and PTHrP [Gardella TJ, 2008]. Depending on its mode of administration, PTH can have either catabolic or anabolic effects on bone. The anabolic actions have been attributed to increased proliferation of osteoprogenitor cells and osteoblasts [Datta et al., 2007; Miao et al., 2002; Pettway et al., 2008], decreased osteoblast apoptosis [Bellido et al., 2003; Chen et al., 2002; Jilka et al., 1999], and/or increased osteoblast differentiation [Yu et al., 2009].
Important insights into mechanisms controlling osteoblast-specific gene expression have been gained through the study of regulatory sequences in the osteoblast-specific gene, osteocalcin. The mouse genome contains two osteocalcin genes, mOG1 and mOG2, having identical patterns of expression and very similar promoter sequences [Ducy and Karsenty, 1995]. The mOG2 promoter contains an OSE1 motif (osteocalcin-specific element 1; TTACATCA) and two OSE2 motifs (AACCACA) within its first 657 bp [Ducy and Karsenty, 1995]. Both motifs are required for osteoblast-specific expression of osteocalcin in vitro and in vivo [Ducy and Karsenty, 1995; Frendo et al., 1998]. The OSE1 core sequence has subsequently been identified as a DNA binding site for the transcription factor ATF4, also known as CREB2 (cAMP-response element-binding protein 2). ATF4 is a basic leucine-zipper transcription factor that is a member of the ATF/CREB protein family.
The crucial role for ATF4 in osteoblast differentiation and bone development was demonstrated using Atf4-deficient mice [Yang et al., 2004b], which exhibit a marked reduction or delay in mineralization of several bones including frontal and parietal bones, clavicles, and long bones. At birth, the delay of mineralization in the skull was still present as demonstrated by widening of the fontanelles. Postnatally, Atf4-deficient mice have a severe reduction in bone volume and in the number and thickness of trabeculae that persists throughout life. ATF4 is required for PTH induction of osteocalcin gene expression in osteoblasts [Yu et al., 2008]. PTH accomplishes this in part by increasing ATF4 mRNA and protein level as well as increasing ATF4 binding to OSE1 DNA. Recently, results from our group demonstrated that ATF4 plays a critical role in the anabolic effects of PTH on bone [Yu et al., 2009].
The OSE2 site of the osteocalcin promoter binds Runx2, a runt domain-containing transcription factor that is essential for osteoblast and hypertrophic chondrocyte differentiation and bone formation during embryogenesis and postnatal life. Runx2 haploinsufficiency causes cleidocranial dysplasia, a disorder characterized by defective endochondral and intramembranous bone formation [Mundlos et al., 1997]. Runx2−/− mice die at birth and lack skeletal ossification and mature osteoblasts [Komori et al., 1997; Mundlos et al., 1997]. During early development, Runx2 is essential for osteoblast differentiation [Ducy et al., 1997]. In terminally differentiated osteoblasts, Runx2 regulates the expression of bone matrix proteins [Ducy et al., 1999]. Runx2 and ATF4 cooperatively regulate osteocalcin transcription through interactions with OSE1 and OSE2 sites in the promoter since mutation of OSE1 markedly reduced Runx2-dependent mOG2 transcription and mutation of either OSE1 or OSE2 dramatically reduced ATF4-stimulated activity [Xiao et al., 2005].
Recent studies identified the basic helix-loop-helix (bHLH)-containing transcription factors, Twist1 and Twist2, as major regulators of Runx2 function. Twist1 is required for closure of the neural tube during mouse development [Chen and Behringer, 1995]; mice homozygous for a Twist2 null allele show elevated expression of proinflammatory cytokines causing perinatal death [Sosic et al., 2003]. Twist proteins transiently inhibit osteoblast differentiation during skeletogenesis through the interaction of a novel domain in these proteins called the Twist box and the Runx2 DNA binding domain [Bialek et al., 2004]. A more recent study suggested that, in addition to the C-terminus Twist box, the bHLH domain of Twist1 also plays a significant role in regulating Runx2 function [Connerney et al., 2006]. In addition, the bHLH domain of Twist1 mediates homo- and heterodimerization with other bHLH family members such that small changes in the level of Twist1 expression alter the ratio of homodimers to heterodimers, which can have dramatic effects on transcriptional regulation.
It was hypothesized that PTH is able to affect osteoblast differentiation by regulating Twist1. We demonstrate that PTH has a profound inhibitory effect on Twist1 mRNA in MC3T3-E1 osteoblasts. Since ATF4 is a major regulator of the PTH response in osteoblasts, studies were extended to explore the interaction between Twist1 and ATF4. As will be shown, Twist proteins inhibit ATF4 binding to OSE1 in part by directly binding to ATF4. This study thus identifies Twist proteins as novel inhibitory binding partners of ATF4 and highlights the functional significance of this interaction.
MATERIALS AND METHODS
Reagents
Tissue culture medium and fetal bovine serum were obtained from Invitrogen (Carlsbad, CA). PTH (1–34, human) was obtained from Bachem (Torrance, CA).
Cell Culture and Cellular Transfections
MC3T3-E1 Subclone 14 cells which exhibit high levels of osteoblast differentiation and mineralization after growth in medium containing ascorbic acid were plated in α-MEM with 10% fetal bovine serum containing 100 U/ml penicillin and 100 mg/ml streptomycin at a density of 50,000 cells/cm2. After 24 h, cells were cultured in medium containing 0.1% FBS in the presence or absence of 10−8 M PTH for 6 hrs. Cultures at passage less than 15 were used for all experiments.
ROS 17/2.8 osteosarcoma cells and 293T human kidney epithelial cells (HEK293T) were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (Invitrogen) and 50 IU/ml penicillin-streptomycin (Mediatech, Herndon, VA). 293T cells were transfected using 25-kDa linear polyethylenimine (Polysciences, Warrington, PA) as described previously [Danciu and Whitman, 2010].
ROS 17/2.8 transfection was performed as follows. Cells were plated at a density of 3×106 cells per 10 cm dish in antibiotic-free medium. Twenty-four hours post plating, cells were transfected using 8.7 μg of pcs4-3flag-mTwist1 DNA, 21.75 μl Lipofectamine LTX and 8.7 μl PLUS reagent (Invitrogen). Forty-eight hours post transfection, cells were stimulated with vehicle or 10−7 M PTH for 6 hrs in complete media.
DNA Constructs
p4OSE1-luc was previously described [Ducy and Karsenty, 1995]. To generate mouse ATF4, we performed PCR with primers containing BglII or XhoI sites using pCMV/ATF4 as template [Yang et al., 2004b]. mATF4 was then subcloned at the BglII/XhoI site of pcs4-3Flag. Plasmids expressing truncated forms of ATF4 (aa 1–333 and aa 1–276) were generated in a similar manner using templates as described previously [Xiao et al., 2005]. One-step site-directed PCR-based mutagenesis was performed using pcs4-3Flag-mATF4 as previously described to generate the following mATF4 deletion mutants: Δ1-90, Δ91-211, Δ1-211, Δ211-276, Δ1-276 [Makarova et al., 2000]. Mouse Twist1 constructs were generated in our laboratory as previously described [Danciu and Whitman, 2010].
Full-length GST-ATF4 fusion protein expression plasmid was constructed by subcloning the full-length ATF4 cDNA into the GST gene fusion vector pGEX-4T-1 (Amersham Biosciences) in correct reading frame as previously described [Xiao et al., 2005]. Full-length HIS-HA-Twist1 was a kind gift of Dr. Gerard Karsenty (Columbia University, NY; [Bialek et al., 2004]) A construct containing full-length human Twist2 was obtained from the Harvard Institute of Proteomics and subcloned using PCR primers containing BglII and EcoRI sites in the pcs4-3Flag vector. One-step site-directed PCR-based mutagenesis was performed using pcs4-3Flag-hTwist2 as a template as described above. All sequences were verified by automatic DNA sequencing.
Quantitative RT-PCR
MC3T3-E1 osteoblasts were plated at a density of 50,000 cells/cm2 in 35 mm dishes and cultured in 10% FBS overnight. Cells were then treated with 10−8M PTH for 6 hrs in 0.1% FBS. RNA was isolated and purified using an RNeasy Mini Kit (Qiagen, Inc., Valencia, CA). Double-stranded complementary DNA (cDNA) was synthesized from 1 μg of RNA, using Oligo d(T) (Applied Biosystems, Foster City, CA) to prime total RNA samples for reverse transcription using Multiscribe reverse transcriptase (Applied Biosystems). The TaqMan universal PCR master mix (Applied Biosystems) was used for detection of cDNA. cDNA was amplified using customized primers and probes (Applied Biosystems). Rodent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. Amplification of cDNA was performed using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Relative quantification of data generated using this system was performed using the standard curve method. Experiments were performed three times with similar results.
Luciferase Assays
p657mOG2-luc, p4OSE1-luc and p657mOG2OSE2mt-luc, which contains 2-bp substitution mutations in the two OSE2 sites at positions −135/−134 and −606/−605 (CCAAGAACA), respectively, were previously described [Xiao et al., 2005]. MC3T3-E1 containing stably integrated copies of a 1.3-kb mOG2 promoter driving a firefly luciferase reporter gene has been previously described by our group [Jiang et al., 2004]. 293T cells were plated in 24 well plates at a density of 2.4 × 105 cells/well and transfected on the same day using PEI as described above with 60 ng p4OSE1-luc, 60 ng pcs4-3HA-mATF4, 60 ng CMV/b-galactosidase (to control for transfection efficiency) and either 60, 120 or 240 ng pcs4-3Flag-mTwist1 or 120 ng pcs4-Twist1-box (mTwist1 missing the C-terminus Twist box). Cells were harvested 48 h after transfection and assayed using a Dual Luciferase assay kit (Promega, Madison, WI) on a Monolight 2010 luminometer (BD Biosciences).
Luciferase assays using MC3T3 osteoblasts were performed as follows. Cells were plated at a density of 50,000 cells/cm2 in 6 well dishes 24 hrs prior to transfection using Lipofectamine LTX (Invitrogen) and 3 μg of p4OSE1-luc and 0.75 μg of CMV/b-galactosidase. 48 hrs after transfection, cells were stimulated with 10−8M PTH in medium containing 0.1% FBS for 6 hrs.
Recombinant proteins and pull-down assays
GST-fusion proteins were isolated from bacterial lysate using glutathione beads, and His-tagged proteins were isolated from bacterial lysate using the Talon system (Clontech) according to manufacturers’ directions. Purified GST was a kind gift of Dr. Jorge Iniguez-Llhui (University of Michigan, MI). For the pull-down, 200 ml of 50% sepharose bead slurry 16 mg of purified GST or GST-ATF4 were incubated for 1hr at 4°C. Purified His-HA-Twist1 protein was precleared using sepharose beads for 1hr at 4°C. Precleared Twist1 protein was then incubated with 20 ml of sepharose-GST or sepharose-GST-ATF4 for 1 hr at 4°C. Five 10 min washes were performed at 4°C as follows: washes 1, 4, 5: 0.1% NP40 in PBS; washes 2, 3: 500 mM NaCl, 0.1% NP40 in PBS.
Western blot analysis and immunoprecipitations
Cultured 293T cells were rinsed twice in ice-cold phosphate-buffered saline then lysed in modified RIPA buffer [(150 mM NaCl, 50 mM Tris (pH 8), 25 mM β-glycerophosphate, 100 mM sodium fluoride, 2 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 2x Complete EDTA-free protease inhibitor mixture (Roche Applied Science), 1 mM phenylmethylsulfonyl fluoride), 2 mM EDTA, 1% Nonidet P-40 (NP-40)]. Lysates were centrifuged and supernatants collected.
Immunoprecipitations were performed using an anti-Flag matrix (Sigma) for 3 hours at 4°C with gentle rocking. Proteins were electrophoretically separated on polyacrylamide gels and then transferred onto nitrocellulose membranes. The following antibodies were used for Western blotting: anti-HA-peroxidase rat monoclonal peroxidase-conjugated antibody (clone 3F10, Roche), anti-Flag-peroxidase mouse monoclonal peroxidase-conjugated antibody (Sigma), and anti-actin mouse monoclonal antibody (Sigma). Rabbit anti-mouse ATF4 antibody for immunoprecipitation and Western blotting was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Flag HRP antibody was from Sigma. Horseradish peroxidase-conjugated goat anti-rabbit or -mouse IgG were from Jackson Immunolabs (West Grove, PA).
Nuclear and cytoplasmic fractions were obtained using the NE-PER extraction kit from Thermo Scientific (Rockford, IL, USA) according to the manufacturer’s directions. Histone3 antibody was obtained form Cell Signaling Technology (Danvers, MA, USA).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed as previously described [Li et al.]. Briefly, ROS 17/2.8 cells transfected with pcs4-3flag-mTwist1 or empty vector as described above, were cross-linked with 1% formaldehyde and sonicated in sonication buffer. The sheared chromatin was then precipitated with ATF4 antibody (Santa Cruz, Biotechnology) or control IgG (anti-rabbit). The immunoprecipited chromatin library was measured by PCR with the following primers: 5—GCAGTCACCAACCACAGCATCCTT—3 (forward) and 5—TCCACTGCCTGAGCCCCTAATGT—3 (reverse). These primers were based on the rat osteocalcin gene sequence flanking a previously identified ATF1 site [Ducy and Karsenty, 1995]. The coding region primers were previously published by our group [Li et al., 2010]. ChIP PCR products were visualized on 2.5% agarose gel and stained with ethidium bromide. Purified input chromatin was used to perform parallel PCR with the respective primer pairs.
Statistical analysis
The differences among groups for the effect of Twist1 on ATF4-mediated OSE1 transcriptional regulation (fig 2A) were statistically assessed by one-way analysis of variation (ANOVA) with Tukey multiple comparison post hoc test using a statistical software package (GraphPad Prism version 4.00, GraphPad Software, San Diego, California). The differences between groups for the effect of PTH stimulation on OSE1- luciferase activity (fig 1A) were statistically assessed by a one-sided Student’s t-test; the differences between groups for PTH-mediated Twist1 regulation as assessed by quantitative PCR were assessed using the Bonferroni test (PASWStatistics 18.0, SPSS Inc., Chicago, IL); fig 1B).
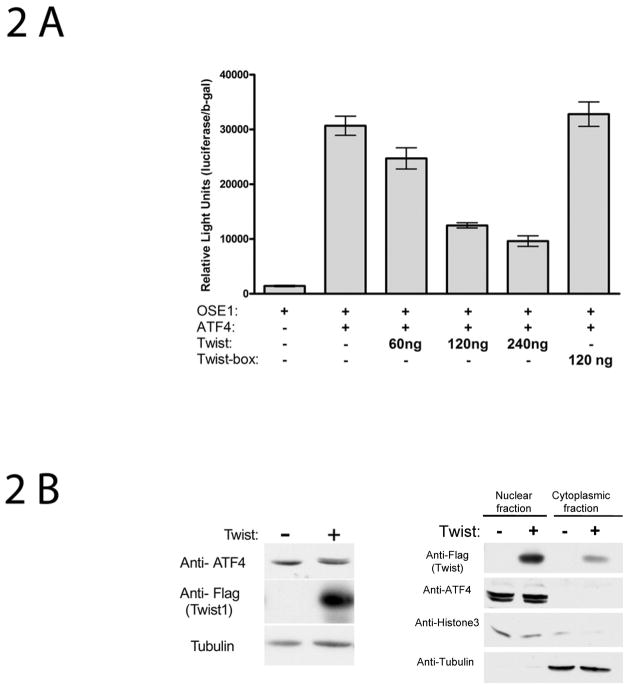
Fig 2.
Twist1 decreases ATF4-dependent transcriptional activity without affecting ATF4 levels. (A) Twist-dependent ATF4 transcriptional regulation in 293T cells—response is dependent upon the C-terminus Twist1 box. Cells were plated in 24 well plates and transiently transfected with 60 ng p4OSE1-luc, 60 ng pcs4-3HA-mATF4, various concentrations of pcs4-3Flag-mTwist1 ranging from 60 ng to 240 ng as indicated, 120 ng pcs4-3Flag-(mTwist1-box) and 60 ng β-galactosidase normalization plasmid. Cells were harvested and assayed for dual-luciferase activity 48 hrs after transfection. Firefly luciferase activity was normalized to b-galactosidase activity (for transfection efficiency). Data represent mean ± SD. Experiments were repeated at least three times and qualitatively identical results were obtained. *: p<0.001.
(B) Left panel, Twist1 overexpression in ROS 17/2.8 osteoblast-like cells does not affect endogenous ATF4 levels. Cells transfected with vector only (pcs4-3Flag) or with Flag-tagged Twist1 were collected 48 h post transfection and processed for Western blotting using the indicated antibodies. Right panel, A significant portion of overexpressed Twist1 protein localizes in the nucleus as does ATF4.
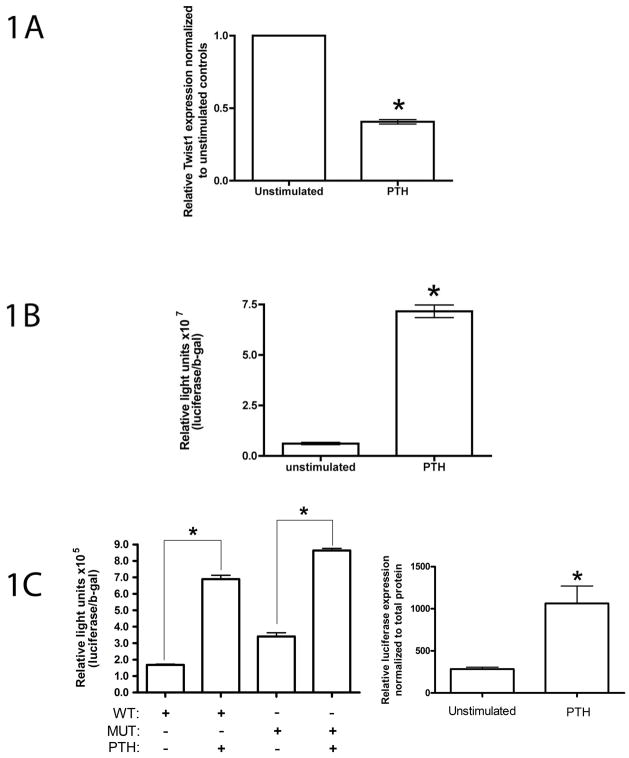
Fig 1.
PTH effects on MC3T3-E1 osteoblasts. Cells were seeded at a density of 50,000 cells/cm2 in 35 mm dishes and cultured in 10% FBS for 24 hrs prior to stimulation with 10−8 M PTH for 6 hrs in 0.1% FBS. (A) PTH stimulation of MC3T3 osteoblasts causes a significant decrease in Twist mRNA. RNA preparation and quantitative real-time RT-PCR for Twist1 was performed and normalized to GAPDH mRNA. Data represent mean ± SD expressed as fold over control. (B) PTH increases ATF4-dependent transcriptional activity in MC3T3-E1 osteoblasts. Cells were transiently transfected with p4OSE-1-luc and β-galactosidase normalization plasmid and treated for 6 hrs with 10−8M PTH in 0.1% FBS. Experiments were repeated at least three times in triplicates. *p<0.0001 (control vs PTH). (C) PTH activates the osteocalcin promoter in MC3T3-E1 osteoblasts. Left panel, cells were transiently transfected with wild-type and mutant p657mOG2-luc (mutations of both OSE2 sites which abolishes the Runx2-dependent induction of promoter activity) and β-galactosidase normalization plasmid and stimulated 48 h post transfection for 6 hrs with 10−8M PTH in 0.1% FBS. Experiments were repeated at least three times in replicates. *p<0.001 (control vs PTH). Right panel, MC3T3-E1 containing stably integrated copies of a 1.3-kb mOG2 promoter driving a firefly luciferase reporter gene were cultured in 10% FBS for 24 hrs prior to stimulation with 10−8 M PTH for 6 hrs in 0.1% FBS. Luciferase activity was normalized to total protein. Experiments were repeated at least three times in replicates. *p<0.001 (control vs PTH).
RESULTS
PTH decreases Twist1 expression in osteoblasts
The observation that Runx2 is expressed in the lateral plate mesoderm at E10 during mouse development [Ducy, 2000] yet expression of molecular markers of differentiated osteoblasts cannot be detected prior to E13 [Bialek et al., 2004], led to the identification of Twist proteins as essential regulators of osteoblast differentiation. Twist proteins do not affect Runx2 expression yet they are able to down-regulate osteocalcin, an osteoblast-specific gene whose expression is under Runx2 control [Bialek et al., 2004]. Because the osteocalcin gene is also regulated by PTH [Boudreaux and Towler, 1996; Yu and Chandrasekhar, 1997], we investigated the effect of PTH stimulation on osteoblast Twist1 transcription. Our group has previously published that the PTH/PTHrP receptor (PPR), plays an important role in the process of proliferation and differentiation of mesenchymal cells into osteoblasts [Chen et al., 2002; McCauley et al., 1995]. Furthermore, our group also has shown that a 6 hr stimulation of MC3T3 osteoblasts with 10−8M PTH resulted in a significant upregulation of osteocalcin mRNA [Jiang et al., 2004]. Following an identical stimulation protocol, we observed at 6 hr an approximately 60% reduction in Twist1 mRNA as assessed by real-time RT-PCR and illustrated in fig 1A. PTH mediates its action on osteocalcin gene expression in part by increasing ATF4 expression and activity in osteoblasts[Jiang et al., 2004]. ATF4 activates osteocalcin transcription through direct binding to the OSE1 site in addition to interactions with Runx2 at the OSE1 and OSE2 sites [Ducy and Karsenty, 1995; Xiao et al., 2005]. We previously demonstrated that an artificial promoter containing four copies of OSE1 fused to a −34 to +13 minimal mOG2 promoter (p4OSE1-luc) was also stimulated by ATF4 in osteoblasts, while the same construct containing mutated OSE1 was not [Xiao et al., 2005]. We confirmed that in our system, 6 hrs of PTH stimulation increased ATF4-dependent transcriptional activity in MC3T3 osteoblasts (fig 1B). Furthermore, following an identical stimulation protocol, we extended our observations to include osteocalcin promoter regulation results. Two osteocalcin promoter regions were tested as follows (fig1C, left and right panels). The 657-bp mOG2 promoter contains a single ATF4-binding site (OSE1) at −48/−55 and two Runx2-binding sites (OSE2) at −131/−137 and −602/−608. Our group has previously demonstrated that mutating both OSE2 sites completely abolishes the Runx2-dependent induction of promoter activity [Xiao et al., 2005]. PTH-activation of the osteocalcin promoter containing mutant OSE2 sites, demonstrates that Runx2 activity (and therefore Twist1 modulation of Runx2 activity) is dispensable for this response. In addition, we used MC3T3-E1 cells containing stably integrated copies of a 1.3-kb mOG2 promoter driving a firefly luciferase reporter gene. In both instances, a 6-h stimulation with 10−8M PTH resulted in a significant activation of osteocalcin transcription. These findings demonstrate that, in our system, PTH decreases Twist1 expression while simultaneously activating osteocalcin transcription.
Twist1 inhibits ATF4 transcriptional activity
Previous time-course experiments in osteoblasts demonstrated that PTH induction of Atf4 occurs within 1 hr of PTH addition and peaks after 3–6 hrs [Yu et al., 2008]. Since PTH increases Atf4 transcription and binding activity and also represses Twist1 transcription over a similar time-course, we hypothesized that Twist1 down-regulation by PTH may be able to further enhance the ATF4 response similar to the way Twist1 downregulation permits cranial suture development by relieving inhibition of Runx2 during development [Bialek et al., 2004]. Therefore, the role of Twist1 in ATF4 regulation of the OSE1 site in the osteocalcin promoter was investigated. As shown in Fig 2A, ATF4 activated p4OSE1-luc and Twist1 was able to inhibit this response in a dose-dependent manner. Furthermore, a Twist1 construct missing the C-terminal domain called “the Twist box” [Bialek et al., 2004] was not able to inhibit ATF4-dependent activation of the luciferase construct. Since the reduction in p4OSE1-luc activity could be due to Twist1 causing a decrease in ATF4 levels, we determined endogenous ATF protein level in ROS osteoblast-like cells overexpressing Twist1. Twist1 overexpression did not affect ATF4 protein levels (fig 2B, left panel). Fig 2B, right panel, demonstrates that overexpressed Twist1 localizes mainly in the nucleus where it is able to interact with ATF4 which is also a nuclear protein.
These results suggest that Twist1 inhibits ATF4 function through mechanisms independent of ATF4 protein level.
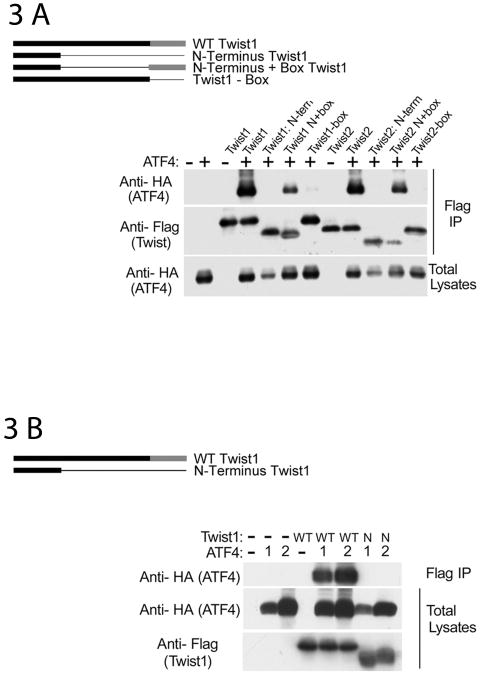
Protein-protein interaction studies reveal a direct interaction between ATF4 and the C-terminal domain of Twist1
To identify the domains within Twist proteins responsible for the inhibition of ATF4 function, we used Twist1 deletion mutants in DNA cotransfection assays. The following Twist1 and Twist2 constructs were used: full-length, N-term (containing the N-terminus only), N+box (missing the bHLH domain), and –box (missing the Twist box) [Bialek et al., 2004]. Figs 3A and B illustrate that both Twist1 and Twist2 interact with ATF4. When Twist constructs missing the bHLH domain were used, the interaction was present but reduced. Twist constructs containing only the N-terminus as well as Twist constructs missing the Twist box failed to interact with ATF4 indicating that the Twist domain necessary for ATF4 interaction is the Twist box, the same domain necessary for Runx2 interaction. To further confirm that ATF4 does not interact with N-term Twist1, we used different concentrations of ATF4 since ATF4 total protein levels are decreased in the presence of N-term Twist1. As fig 3B indicates, N-term Twist1 fails to interact with ATF4 even at high ATF4 protein levels.
Fig 3.
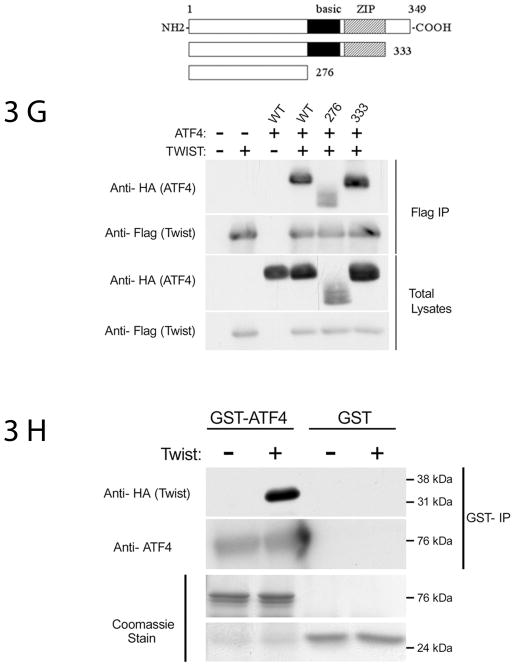
ATF4 and Twist interact directly—mapping of the domains necessary for this interaction. (A) Twist C-terminal box domain is necessary and sufficient for Twist1 and Twist2 interaction with ATF4. 293T cells were transfected with the following constructs in pcs4-3Flag vector: mTwist1, Twist1 N-term (mTwist1 containing the N-terminus only), Twist1 N+box (mTwist1 missing the bHLH domain), Twist1-box (mTwist1 missing the C-terminus Twist box); similar constructs were generated using Twist2. Cells were also transfected with an equal amounts of pcs4-3HA-mATF4 as indicated. Upper panels are of the flag immunoprecipitations blotted with anti-HA antibody or with an anti-Flag antibody to detect ATF4 or Twist respectively. An anti-HA Western blot of whole lysates indicates that ATF4 level was decreased in the presence of Twist1 or Twist2 N-term. (B) ATF4 does not interact with Twist1 N-term (which does not have the bHLH and the box domains). 293T cells were transfected with either 0.5 μg (denoted as “1”) or 1.5 μg (denoted as “2) of pcs4-3HA-mATF4 in the presence or absence of 0.5 μg WT or mutant (Twist1 N-term) pcs4-3Flag-mTwist1. Flag immunoprecipitations were blotted with an anti-HA antibody to detect ATF4; total lysates were blotted with anti-HA and anti-Flag antibodies to detect ATF4 and Twist1 levels, respectively. (3) Twist1 interacts with aa 1-276 of ATF4. 293T cells were transfected with pcs4-3Flag-mTwist1 and an equal amount of pcs4-3HA-mATF4 or Δ1-276mATF4 (mutant missing aa 1-276). Flag immunoprecipitations were blotted with an anti-HA antibody to detect ATF4; total lysates were blotted with anti-HA and anti-Flag antibodies to detect ATF4 and Twist1 levels, respectively. (D,E) Twist1 interacts with various ATF4 mutants containing partial deletions of aa 1-276. 293T cells were transfected with 0.5 μg pcs4-3Flag-mTwist1 and an equal amount of pcs4-3HA-mATF4 or the various ATF4 mutants (Δ1-90: missing the first 90 aa; Δ91-211: missing aa 91-211; Δ211-276: missing aa 211-276). (F) Due to the lower levels of total ATF4 protein observed when the Δ1-90 mutant was used, two different concentrations of pcs4-3HA-Δ1-90ATF4 were used: 0.5 μg and 1.5 μg as illustrated in the lower right panel. Lower left panel also demonstrates that aa 91 to 276 are dispensable for Twist1 interaction. For all panels, Flag immunoprecipitations were blotted with an anti-HA antibody to detect ATF4; total lysates were blotted with anti-HA and anti-Flag antibodies to detect ATF4 and Twist1 levels, respectively. (G) The bZIP and the carboxy-terminal domains of ATF4 are dispensable for Twist1 interaction. 293T cells were transfected with pcs4-3Flag-mTwist1 and an equal amount of pcs4-3HA-mATF4, 1-276mATF4 (mutant containing a stop codon after aa 276), or 1-333mATF4 (mutant containing a stop codon after aa 333). Flag immunoprecipitations were blotted with an anti-HA antibody to detect ATF4; total lysates were blotted with anti-HA and anti-Flag antibodies to detect ATF4 and Twist1 levels, respectively. (H) Twist1 and ATF4 interact directly (pull-down assays). GST-ATF4 or GST alone complexed to glutathione beads were incubated with His-HA-Twist-1. Eluates from the beads were immunoblotted with anti-HA antibody to detect Twist1 and ATF4 antibody. Only GST-ATF4 and His-Twist1 showed direct interaction. Lower panels show equal loading of GST-ATF4 and GST as indicated by Coomassie staining of the gel.
To map the ATF4 domain necessary for the Twist-ATF4 interaction, we overexpressed Flag-tagged Twist1 and various HA-tagged ATF4 constructs. Total lysates were immunoprecipitated with an anti-Flag matrix, followed by Western blot analysis using an anti-HA antibody. As shown in figs 3C-F, ATF4 protein was present in an anti-Twist1 immunoprecipitate. Twist1 was not immunoprecipitated by anti-ATF4 antibodies in the absence of ATF4. Fig 3G illustrates that deletion of the basic region and the whole leucine zipper domain (ATF4 containing the first 276 aa) did not abolish ATF4 interaction with Twist1; the decrease in interaction reflects lower total ATF4 protein level of the 276 (stop) ATF4 construct. An ATF4 construct missing the first 211 aa (Δ1-211) or the first 276 aa (Δ1-276) is not able to interact with Twist1 suggesting that the first 211 aa of ATF4 are necessary for its interaction with Twist1 (figs 3C and D). To characterize this region further, we used two ATF4 deletion mutants: ATF4 construct missing the first 90 aa (Δ1-90), and aa 91-211 (Δ91-211). As figs 3D and E illustrate, Twist1 interacts with both deletion mutants of ATF4. This finding suggests that while the first 211 aa of ATF4 are necessary for Twist1 interaction, a complex interaction pattern might occur whereby multiple motifs within this region are involved in protein-protein interactions. Furthermore, fig 3G illustrates that the ATF4 domain interacting with Twist1 is located between aa 1 and 276 and that the basic, ZIP and the carboxy-terminal domains are dispensable for this interaction.
To determine whether ATF4 and Twist1 can directly interact in the absence of other nuclear proteins, we conducted standard GST pull-down assays using purified full-length GST-ATF4 and HIS-Twist1 fusion proteins. Consistent with our immunoprecipitation results using total cell lysates from cells transfected with ATF4 and Twist1, purified HIS-Twist1 was complexed to GST-ATF4 but not to GST alone (fig 3D). The lower panels in fig 3D are of Coomassie-stained gel and demonstrate equal levels of GST and GST-ATF4 protein levels.
Twist1 overexpression in osteoblasts prevents ATF4 binding to the osteocalcin OSE1 site in response to PTH
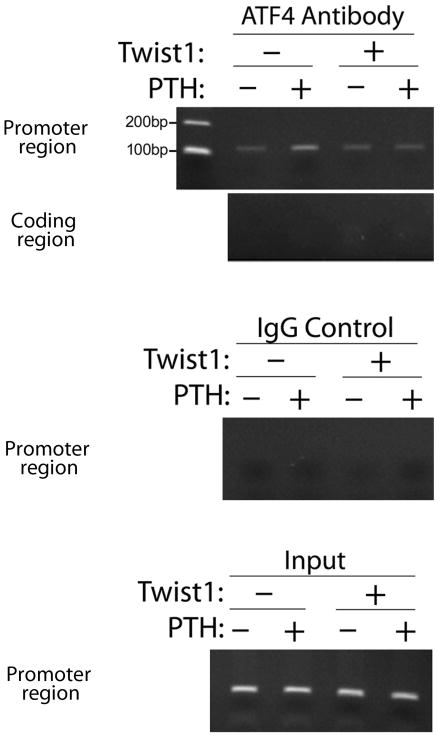
The ROS cell line derived from a rat osteosarcoma, is a widely used cell line to study osteoblast-specific gene expression [Ducy and Karsenty, 1995]. ROS 17/2.8 cells have markers of both immature and more differentiated osteoblasts [McCabe et al., 1994] and most importantly for this study they have highly functional parathyroid hormone receptors. Furthermore, these cells do not express Twist1 as assessed by Northern blotting [Bialek et al., 2004]. To determine the effect of overexpressed Twist1 on the PTH-induced association of ATF4 with the endogenous osteocalcin promoter in osteoblasts, we performed chromatin immunoprecipitation (ChIP) assays. For this assay we used ROS cells overexpressing Twist1 or vector only, with or without PTH treatment. Figure 2B illustrates that in these cells, Twist1 overexpression does not affect total ATF4 levels. As shown in fig 4, ATF4 interacted specifically with a chromatin fragment of the proximal osteocalcin promoter that contains the ATF4-binding site (OSE1); this interaction was dramatically stimulated by PTH treatment as we previously reported [Yu et al., 2008]. Moreover, PTH treatment of cells overexpressing Twist1 did not stimulate ATF4 binding to the OSE1 site in the osteocalcin promoter supporting our hypothesis that Twist1 is a negative regulator of ATF4 function.
Fig 4.
Twist1 overexpression attenuates PTH-induced binding of endogenous ATF4 to the OSE1 site of the osteocalcin promoter in ROS 17/2.8 osteoblast-like cells. ROS cells transfected with pcs4-3flag-mTwist1 or vector only control (pcs4-3flag) were stimulated with 10−7M PTH for 6 hrs as detailed in materials and methods. A ChIP assay was then performed of the rat osteocalcin promoter region that contains the ATF-4 binding site OSE1. Two negative controls were used (ATF4 immunoprecipitate analyzed with coding region primers and IgG control immunoprecipitate analyzed with promoter region primers).
DISCUSSION
Our finding that Twist1 interacts directly with ATF4 and that this interaction inhibits PTH-induced binding of ATF4 to the osteocalcin promoter, further extends the critical role of Twist1 in regulating osteoblast function. In addition to increasing osteoblast cell number by stimulating osteoblast proliferation and decreasing osteoblast apoptosis, intermittent PTH also stimulates osteoblast differentiation [Iida-Klein et al., 2002; Schmidt et al., 1995; Wang et al., 2005], an effect that is in part mediated by ATF4 [Yu et al., 2009]. In vivo, PTH elevates the expression of genes known to be associated with osteoblast differentiation including: osteocalcin, bone sialoprotein, alkaline phosphatase, osteopontin, and osterix. This effect on gene expression is either reduced or completely abolished in ATF4−/− mice [Yu et al., 2009], thus the identification of novel regulators of ATF4 function may provide important insights into the mechanisms for the anabolic actions of PTH on bone.
Twist proteins have recently been identified as negative regulators of osteoblast differentiation. Both Twist1 and Twist2 have a conserved C-terminus domain called the Twist box that interacts with Runx2 and inhibits the expression of downstream targets of Runx2 including osteocalcin, an osteoblast-specific gene [Bialek et al., 2004]. Osteocalcin transcription is regulated through OSE1 and OSE2 core sequences in its promoter. Although ATF4 binds to OSE1 and uses this site to activate the osteocalcin promoter [Yang et al., 2004b], Runx2 is also necessary for this activation as ATF4 is not able to induce osteocalcin mRNA expression in non-osteoblastic cells that lack detectable Runx2 [Xiao et al., 2005]. A further demonstration of this cooperativity between Runx2 and ATF4 is illustrated by studies in which mutation of either OSE1 or OSE2 dramatically reduced ATF4-stimulated Runx2 activity [Xiao et al., 2005]. The relatively rapid decrease of Twist1 mRNA seen after PTH treatment suggests that, in addition to stimulating ATF4 level and activity [Yu et al., 2008], PTH promotes transcription by decreasing Twist1, an inhibitor of both Runx2 and ATF4. In a manner similar to Twist1 regulation of Runx2, Twist1 inhibits ATF4 function without affecting ATF4 protein levels. bHLH and CREB family interactions have been previously reported in the context of muscle and testis development [Muir et al., 2008] where it was demonstrated that the bHLH domain of scleraxis or paraxis was sufficient for interaction with the C-terminal region of ATF4. Although Twist family members were not tested in that study, the finding of a bZIP/bHLH interaction is in contrast to ours demonstrating that the bHLH domain of Twist is dispensable for ATF4 binding. Paraxis, scleraxis, Twist1 and other bHLH family members share a high degree of homology in the bHLH domain [Burgess et al., 1995] so we expected the interaction between Twist1 and ATF4 to also depend on the bZIP/bHLH domains. However, our data showing that the N+box Twist1 (Twist1 mutant missing the entire bHLH domain) or the corresponding Twist2 mutant interact with ATF4 clearly demonstrate that the bHLH domain of Twist is not involved in this interaction. The box domains of Twist1 and Twist2 are highly conserved across species and represent a unique feature for these two bHLH domain family members. In addition to a direct interaction between bHLH and CREB/ATF proteins, indirect interactions mediated by adaptor/linker proteins such as CREB binding protein (CBP/P300) that have the ability to bind both bHLH and bZIP domain proteins, have also been demonstrated [Chaudhary and Skinner, 2001]. Our finding of an interaction between the box domain of Twist proteins with a non-bZIP region of ATF4 adds to the complexity of interactions between these two families of transcription factors.
In addition to Runx2, osterix (Osx) is another transcription factor essential for osteoblast differentiation and bone formation [Nakashima et al., 2002]. We have shown that intermittent PTH increases in vivo Osx expression through an ATF4-dependent pathway [Yu et al., 2009]. Results presented in this study showing that Twist1 interacts with and inhibits the function of ATF4, suggest that Twist1 may have broad effects on PTH-mediated osteoblast differentiation that include Osx in addition to osteocalcin transcription. ATF4 deficiency reduces, but does not completely block the anabolic actions of PTH. Ovariectomized growing and adult ATF4−/− mice still show a modest increase in bone volume in response to PTH [Yu et al., 2009] suggesting that other factors in addition to ATF4 must be involved in the PTH response. In particular, involvement of Runx2 in the anabolic actions of PTH has been suggested [Bellido et al., 2003; Krishnan et al., 2003]. Since Twist is also a negative regulator of Runx2, our studies may have broader significance related to the overall control of osteoblast gene expression by PTH via Twist inhibition of multiple transcription factors.
Although it was initially hypothesized that the bHLH domain of Twist proteins is dispensable for their anti-osteogenic function [Bialek et al., 2004], recent data reveals a more complex pattern of osteoblast regulation by these proteins. Studies from the Spicer laboratory, demonstrated that partner selection of Twist1 has functional consequences as Twist1 homodimers regulate different genes compared to Twist1 heterodimers (Twist1/other bHLH protein) [Connerney et al., 2006]. More importantly, Twist1 requires the presence of its bHLH family member E2A E12 to inhibit the differentiation of calvarial cells. Very little is known about how interactions through the bHLH domain affect C-terminal Twist box interactions and vice versa; further studies are required to determine if the Twist1-ATF4 interaction is affected by dimerization through the HLH domain. Interestingly, we previously demonstrated that disulfide bond formation between Twist1 homodimers significantly reduced its ability to interact with Runx2, indicating that disulfide dimerization in response to an oxidative stress has functional significance [Danciu and Whitman, 2010]. Since Twist1 interaction with ATF4 is mediated through the same C-terminal domain that is involved in Runx2 binding, it is likely that oxidative stress as occurs during prolonged states of inflammation, would alter ATF4-dependent PTH responsiveness of osteoblasts.
In addition to their effects on bone, Twist family members are major regulators of epithelial-to-mesenchymal transition (EMT) during development and carcinogenesis. EMT is a process by which epithelial cells lose their polarity and are converted to a mesenchymal phenotype. One of the characteristic findings in EMT is loss of E-cadherin expression leading to tumor progression, metastasis, and poor prognosis in various human carcinomas; one possible mechanism involved in Twist-mediated EMT is repression of E-cadherin [Yang et al., 2004a]. An intriguing recent finding also identifies ATF4 as required for regulation of EMT [Suzuki et al.] in the context of neural crest development. The role of ATF4 in carcinogenesis and the regulation of EMT by Twist and ATF4 are topics of future studies in our laboratory. PTHrP is produced by a majority of tumors that metastasize to bone and its expression correlates with skeletal localization of tumors [Liao and McCauley, 2006]. A recent study has implicated PTHrP in EMT contributing to renal fibosis [Ardura et al., 2010]. Whether PTHrP plays a role during tumor-associated EMT and how PTHrP affects essential regulators of EMT such as Twist1, is currently unknown.
In conclusion, this study provides the first evidence that Twist1 and Twist2 modulate ATF4-dependent transcriptional activity in response to PTH. Further studies aimed at identifying how PTH regulates Twist transcription will improve our understanding of how this hormone controls osteoblasts and bone remodeling.
Acknowledgments
Grant information:
NIH-NIDCR K22DE016614 to T.E.D.; NIH-NIDDK DK072230 to G.X.; NIH-NIDDK DK 53904 to L.K.M.; NIH-NIDCR DE11723 and DE13386 to R.T.F.
We thank Jennifer Fox for technical assistance and preparation of figures. We are grateful to Jan Berry for valuable comments and technical advice. We thank the H. Rios and W. Giannobile labs for assistance with statistical analysis.
Footnotes
CONFLICT OF INTEREST
None.
LITERATURE CITED
- Ardura JA, Rayego-Mateos S, Ramila D, Ruiz-Ortega M, Esbrit P. Parathyroid hormone-related protein promotes epithelial-mesenchymal transition. J Am Soc Nephrol. 2010;21:237–48. doi: 10.1681/ASN.2009050462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellido T, Ali AA, Plotkin LI, Fu Q, Gubrij I, Roberson PK, Weinstein RS, O’Brien CA, Manolagas SC, Jilka RL. Proteasomal degradation of Runx2 shortens parathyroid hormone-induced anti-apoptotic signaling in osteoblasts. A putative explanation for why intermittent administration is needed for bone anabolism. J Biol Chem. 2003;278:50259–72. doi: 10.1074/jbc.M307444200. [DOI] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–35. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Boudreaux JM, Towler DA. Synergistic induction of osteocalcin gene expression: identification of a bipartite element conferring fibroblast growth factor 2 and cyclic AMP responsiveness in the rat osteocalcin promoter. J Biol Chem. 1996;271:7508–15. doi: 10.1074/jbc.271.13.7508. [DOI] [PubMed] [Google Scholar]
- Burgess R, Cserjesi P, Ligon KL, Olson EN. Paraxis: a basic helix-loop-helix protein expressed in paraxial mesoderm and developing somites. Dev Biol. 1995;168:296–306. doi: 10.1006/dbio.1995.1081. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK. Role of the transcriptional coactivator CBP/p300 in linking basic helix-loop-helix and CREB responses for follicle-stimulating hormone-mediated activation of the transferrin promoter in Sertoli cells. Biol Reprod. 2001;65:568–74. doi: 10.1095/biolreprod65.2.568. [DOI] [PubMed] [Google Scholar]
- Chen HL, Demiralp B, Schneider A, Koh AJ, Silve C, Wang CY, McCauley LK. Parathyroid hormone and parathyroid hormone-related protein exert both pro- and anti-apoptotic effects in mesenchymal cells. J Biol Chem. 2002;277:19374–81. doi: 10.1074/jbc.M108913200. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–99. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–57. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- Danciu TE, Whitman M. Oxidative stress drives disulfide bond formation between basic helix-loop-helix transcription factors. J Cell Biochem. 2010;109:417–24. doi: 10.1002/jcb.22415. [DOI] [PubMed] [Google Scholar]
- Datta NS, Pettway GJ, Chen C, Koh AJ, McCauley LK. Cyclin D1 as a target for the proliferative effects of PTH and PTHrP in early osteoblastic cells. J Bone Miner Res. 2007;22:951–64. doi: 10.1359/jbmr.070328. [DOI] [PubMed] [Google Scholar]
- Ducy P. Cbfa1: a molecular switch in osteoblast biology. Dev Dyn. 2000;219:461–71. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1074>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–36. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Frendo JL, Xiao G, Fuchs S, Franceschi RT, Karsenty G, Ducy P. Functional hierarchy between two OSE2 elements in the control of osteocalcin gene expression in vivo. J Biol Chem. 1998;273:30509–16. doi: 10.1074/jbc.273.46.30509. [DOI] [PubMed] [Google Scholar]
- Gardella TJBF, Potts JT. Receptors for Parathyroid Hormone (PTH) and PTH-related protein. Academic Press, Inc; 2008. [Google Scholar]
- Iida-Klein A, Zhou H, Lu SS, Levine LR, Ducayen-Knowles M, Dempster DW, Nieves J, Lindsay R. Anabolic action of parathyroid hormone is skeletal site specific at the tissue and cellular levels in mice. J Bone Miner Res. 2002;17:808–16. doi: 10.1359/jbmr.2002.17.5.808. [DOI] [PubMed] [Google Scholar]
- Jiang D, Franceschi RT, Boules H, Xiao G. Parathyroid hormone induction of the osteocalcin gene. Requirement for an osteoblast-specific element 1 sequence in the promoter and involvement of multiple-signaling pathways. J Biol Chem. 2004;279:5329–37. doi: 10.1074/jbc.M311547200. [DOI] [PubMed] [Google Scholar]
- Jilka RL, Weinstein RS, Bellido T, Roberson P, Parfitt AM, Manolagas SC. Increased bone formation by prevention of osteoblast apoptosis with parathyroid hormone. J Clin Invest. 1999;104:439–46. doi: 10.1172/JCI6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Moore TL, Ma YL, Helvering LM, Frolik CA, Valasek KM, Ducy P, Geiser AG. Parathyroid hormone bone anabolic action requires Cbfa1/Runx2-dependent signaling. Mol Endocrinol. 2003;17:423–35. doi: 10.1210/me.2002-0225. [DOI] [PubMed] [Google Scholar]
- Li Y, Ge C, Franceschi RT. Differentiation-dependent association of phosphorylated extracellular signal-regulated kinase with the chromatin of osteoblast-related genes. J Bone Miner Res. 25:154–63. doi: 10.1359/jbmr.090705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ge C, Franceschi RT. Differentiation-dependent association of phosphorylated extracellular signal-regulated kinase with the chromatin of osteoblast-related genes. J Bone Miner Res. 2010;25:154–63. doi: 10.1359/jbmr.090705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, McCauley LK. Skeletal metastasis: Established and emerging roles of parathyroid hormone related protein (PTHrP) Cancer Metastasis Rev. 2006;25:559–71. doi: 10.1007/s10555-006-9033-z. [DOI] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques. 2000;29:970–2. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- McCabe LR, Last TJ, Lynch M, Lian J, Stein J, Stein G. Expression of cell growth and bone phenotypic genes during the cell cycle of normal diploid osteoblasts and osteosarcoma cells. J Cell Biochem. 1994;56:274–82. doi: 10.1002/jcb.240560221. [DOI] [PubMed] [Google Scholar]
- McCauley LK, Koh AJ, Beecher CA, Cui Y, Decker JD, Franceschi RT. Effects of differentiation and transforming growth factor beta 1 on PTH/PTHrP receptor mRNA levels in MC3T3-E1 cells. J Bone Miner Res. 1995;10:1243–55. doi: 10.1002/jbmr.5650100815. [DOI] [PubMed] [Google Scholar]
- Miao D, He B, Karaplis AC, Goltzman D. Parathyroid hormone is essential for normal fetal bone formation. J Clin Invest. 2002;109:1173–82. doi: 10.1172/JCI14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir T, Wilson-Rawls J, Stevens JD, Rawls A, Schweitzer R, Kang C, Skinner MK. Integration of CREB and bHLH transcriptional signaling pathways through direct heterodimerization of the proteins: role in muscle and testis development. Mol Reprod Dev. 2008;75:1637–52. doi: 10.1002/mrd.20902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–9. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Pettway GJ, Meganck JA, Koh AJ, Keller ET, Goldstein SA, McCauley LK. Parathyroid hormone mediates bone growth through the regulation of osteoblast proliferation and differentiation. Bone. 2008;42:806–18. doi: 10.1016/j.bone.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt IU, Dobnig H, Turner RT. Intermittent parathyroid hormone treatment increases osteoblast number, steady state messenger ribonucleic acid levels for osteocalcin, and bone formation in tibial metaphysis of hypophysectomized female rats. Endocrinology. 1995;136:5127–34. doi: 10.1210/endo.136.11.7588250. [DOI] [PubMed] [Google Scholar]
- Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–80. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Osumi N, Wakamatsu Y. Stabilization of ATF4 protein is required for the regulation of epithelial-mesenchymal transition of the avian neural crest. Dev Biol. doi: 10.1016/j.ydbio.2010.05.492. [DOI] [PubMed] [Google Scholar]
- Wang YH, Liu Y, Buhl K, Rowe DW. Comparison of the action of transient and continuous PTH on primary osteoblast cultures expressing differentiation stage-specific GFP. J Bone Miner Res. 2005;20:5–14. doi: 10.1359/JBMR.041016. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem. 2005;280:30689–96. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004a;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004b;117:387–98. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Yu S, Franceschi RT, Luo M, Fan J, Jiang D, Cao H, Kwon TG, Lai Y, Zhang J, Patrene K, Hankenson K, Roodman GD, Xiao G. Critical role of activating transcription factor 4 in the anabolic actions of parathyroid hormone in bone. PLoS One. 2009;4:e7583. doi: 10.1371/journal.pone.0007583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Franceschi RT, Luo M, Zhang X, Jiang D, Lai Y, Jiang Y, Zhang J, Xiao G. Parathyroid hormone increases activating transcription factor 4 expression and activity in osteoblasts: requirement for osteocalcin gene expression. Endocrinology. 2008;149:1960–8. doi: 10.1210/en.2007-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XP, Chandrasekhar S. Parathyroid hormone (PTH 1–34) regulation of rat osteocalcin gene transcription. Endocrinology. 1997;138:3085–92. doi: 10.1210/endo.138.8.5315. [DOI] [PubMed] [Google Scholar]