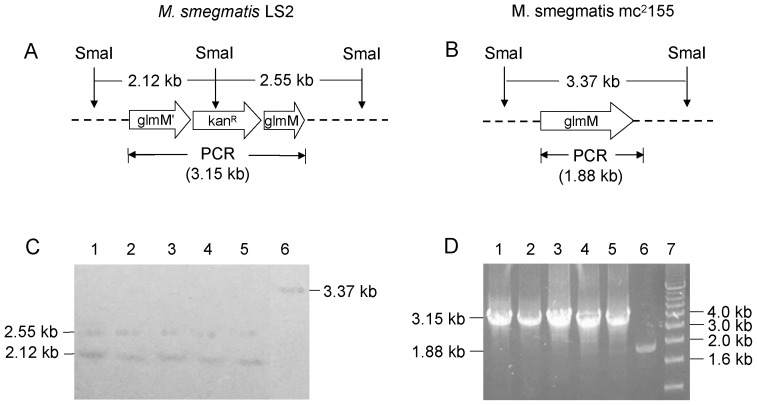
Figure 5. Southern blot and PCR analyses of M. smegmatis LS2 strains.
The genomic DNA was digested overnight by SmaI enzyme. The resulting DNA fragments were separated by a 0.8% agarose gel. The DNA was then transferred to Hybond-N+ membrane and hybridized by MSMEG_1556 probe. A. The expected DNA fragments hybridized by MSMEG_1556 probe were 2.12 kb and 2.55 kb. The expected size of PCR product from M. smegmatis LS2 strain was 3.15 kb. B. The expected DNA fragments hybridized by MSMEG_1556 probe was 3.37 kb and the expected size of PCR product from M. smegmatis mc2155 strain was 1.88 kb. C. Confirmation of M. smegmatis LS2 strains by Southern blot. Lanes 1–5. M. smegmatis LS2 strains have the hybridized DNA bands of 2.12 kb and 2.55 kb; lane 6. wild type M. smegmatis mc2155 has the hybridized DNA band of 3.37 kb. D. Confirmation of M. smegmatis LS2 strains by PCR. Lanes 1–5. the PCR product (3.15 kb) from M. smegmatis LS2 strains; lane 6. the PCR product (1.88 kb) from wild type M. smegmatis mc2155.