Figure 2.
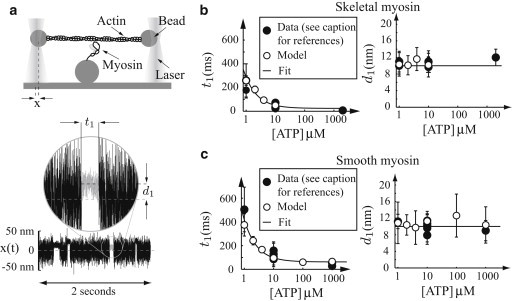
Model is consistent with single-molecule measurements in the laser trap. In all plots: (open circles) Simulated data; (solid circles) experimental data. (Error bars) Standard deviations. (a) A cartoon of a single-molecule experiment. Actin, manipulated by two beads trapped in lasers, is brought close to a myosin molecule that can then bind. The position of one laser-trapped bead x(t) is measured with a quadrant photodiode as a function of time, and the duration of binding (t1) and myosin’s step size (d1) may be measured. A typical simulated experiment is pictured (compare to Fig. 2 of Baker et al. (11)). (b and c) Same plots for skeletal muscle and smooth muscle myosin, respectively. (Left plot) Strong binding lifetime as a function of ATP. The simulated data are fit with an equation of the form t1 = 1/ + 1/kt[ATP] (solid line). (Right plot) Unitary step size d1 as a function of ATP. (Solid line) d1 = 10 nm. Data are from the literature (5,6,8,39,40,43) for smooth muscle and the literature (2,5,37,41,42) for skeletal muscle myosin.
