Figure 3.
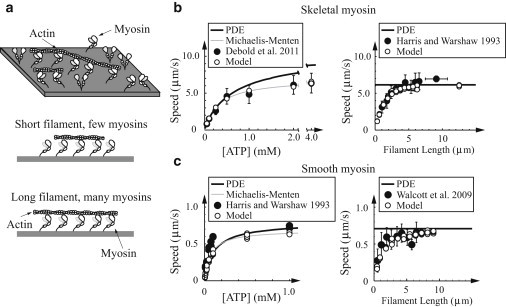
Model is consistent with small and large ensemble measurements in the absence of external force. (a) Cartoon of experimental setup. (Top) Motility assay, with free actin filaments moving across a bed of myosin. (Bottom) Number of myosin molecules interacting with actin depends on filament length. (b and c) Same plots for skeletal and smooth muscle myosin, respectively. In all plots: (open circles) simulated data; (solid circles) experimental data. (Error bars, when present) Standard deviations. (Solid) Numerical solution of the integro-partial differential equations (PDEs) that govern the model in the limit of large N. (Left) In vitro motility at high myosin density as a function of ATP. (Shaded line) Michaelis-Menten fits. Skeletal muscle simulations agree with the experimental measurements (45), given N = 50. The Km of the smooth muscle fits differ from measurements (44) (70.4 μM and 46 μM, respectively). This difference might be due to temperature differences or experimental variation. (Right) In vitro motility speed at low myosin density as a function of filament length. The model is consistent with the data of Harris and Warshaw (44), given 16 active myosin heads/μm (skeletal) and Walcott et al. (43), given 11 active myosin heads/μm (smooth).
