Abstract
Molecules of the title compound, C18H16O6, are almost planar [maximum deviation = 0.096 (4) Å] and reside on crystallographic centres of inversion. They adopt a conformation in which the Cmethyl—O bonds are directed along the molecular short axis [C—C—O—C torsion angles of −175.3 (3) and 178.2 (3)°]. In the crystal, molecules adopt a slipped-parallel arrangement with π–π stacking interactions along the a axis with an interplanar distance of 3.392 (4) Å. Weak C—H⋯O interactions link the molecules into sheets parallel to (10-2).
Related literature
For a study of the effects of alkoxy substituents on the structures and solid-state photophysics, see: Ohta et al. (2012 ▶). For the synthesis, see: Boldt (1967 ▶). For a related structure, see: Kitamura et al. (2009 ▶).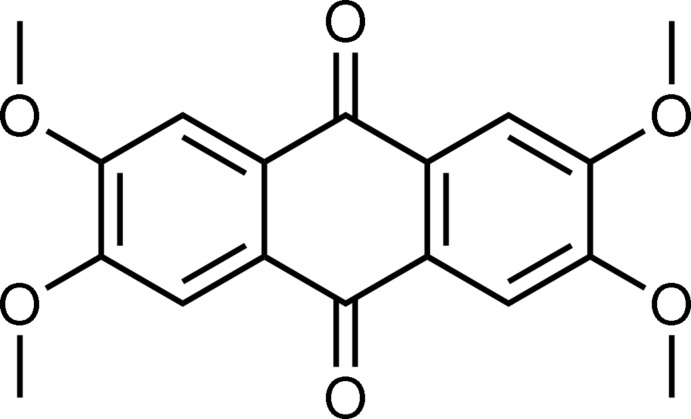
Experimental
Crystal data
C18H16O6
M r = 328.31
Triclinic,

a = 4.6607 (4) Å
b = 8.4769 (9) Å
c = 9.8110 (9) Å
α = 94.859 (3)°
β = 91.410 (2)°
γ = 97.278 (2)°
V = 382.87 (6) Å3
Z = 1
Mo Kα radiation
μ = 0.11 mm−1
T = 223 K
0.50 × 0.06 × 0.05 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
3738 measured reflections
1725 independent reflections
977 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.054
wR(F 2) = 0.234
S = 1.16
1725 reflections
111 parameters
H-atom parameters constrained
Δρmax = 0.39 e Å−3
Δρmin = −0.44 e Å−3
Data collection: RAPID-AUTO (Rigaku, 1999 ▶); cell refinement: PROCESS-AUTO; data reduction: PROCESS-AUTO; program(s) used to solve structure: SIR2004 (Burla et al., 2005 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812033119/gg2091sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812033119/gg2091Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812033119/gg2091Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C8—H8A⋯O1i | 0.97 | 2.58 | 3.391 (5) | 142 |
| C9—H9B⋯O2ii | 0.97 | 2.54 | 3.494 (4) | 168 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was partly supported by a Grant-in-Aid for Scientific Research (C) (No. 23550161) from JSPS and a Grant-in-Aid for Scientific Research on Innovative Areas (No. 23108720, "pi-Space") from MEXT.
supplementary crystallographic information
Comment
9,10-Anthraquinone is an important molecule in the field of industrial dyes. We have recently been interested in the tuning of the solid-state optical properties by the introduction of substituents. As part of our program aimed at the elucidation of the effects of alkoxy substituents on the optical properties in the solid state (Ohta, et al., 2012), we are in need of the information on the crystal structures of a variety of 2,3,6,7-tetraalkoxy-9,10-anthraquinones in order to clarify the correlation between crystal structures and the solid-state photophysics. Although the title compound is already known (Boldt, 1967), the X-ray structure was not reported to date. We report herein the crystal structure of the title compound (I).
The molecular structure of the title compound (I) is shown in Fig. 1. The molecule possesses a center of inversion, and half of the formula unit is crystallographically independent. The molecule is almost planar with the maximum deviation of 0.096 (4) Å for C8. The displacements of atoms O1, O2, O3, C8 and C9 relative to the plane of the anthraquinone framework are 0.023 (2), -0.002 (2), 0.013 (2), 0.096 (4), and 0.060 (3), respectively. The molecule prefers the conformation in which the Cmethyl—O bonds are directed along the molecular short axis. The torsion angles of C3—C2—O2—C8 and C2—C3—O3—C9 are -175.3 (3) and 178.2 (3)°, respectively. This conformation is similar to the corresponding moiety in 2,3-dimethoxy-5,12-tetracenequinone (Kitamura, et al., 2009). The molecules adopt a slipped-parallel arrangement as shown in Fig. 2. Then molecules are π-stacked along the a axis with an interplanar distance 3.392 (4) Å.
To examine the influence of crystal packing on the solid-state fluorescences, the fluorescence spectrum and the absolute quantum yield of (I) were measured with a Hamamatsu Photonics PMA11 calibrated optical multichannel analyzer with a solid-state blue laser (λex = 377 nm) and a Labsphere IS IS-040-SF integrating sphere, respectively. The crystals showed negligible fluorescence (Φ = 0.002). The fluorescence quenching would be due to the π-stacked structure.
Experimental
The title compound was prepared according to the literature procedure (Boldt, 1967). Single crystals suitable for X-ray analysis were obtained by recrystallization from DMF.
Refinement
All the H atoms were positioned geometrically and refined using a riding model with C—H = 0.94 Å and Uiso(H) = 1.2Ueq(C) for aromatic C—H, and C—H = 0.97 Å and Uiso(H) = 1.5Ueq(C) for CH3. The positions of methyl H atoms were optimized rotationally.
Figures
Fig. 1.
The molecular structure of the title compound (I), showing the atomic numbering numbering and 30% probability displacement ellipsoids. Symmetry code: (i) -x, -y + 1, -z.
Fig. 2.
The packing diagram of (I). Hydrogen atoms are omitted for clarity.
Crystal data
| C18H16O6 | Z = 1 |
| Mr = 328.31 | F(000) = 172 |
| Triclinic, P1 | Dx = 1.424 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.6607 (4) Å | Cell parameters from 1977 reflections |
| b = 8.4769 (9) Å | θ = 3.1–27.5° |
| c = 9.8110 (9) Å | µ = 0.11 mm−1 |
| α = 94.859 (3)° | T = 223 K |
| β = 91.410 (2)° | Needle, yellow |
| γ = 97.278 (2)° | 0.50 × 0.06 × 0.05 mm |
| V = 382.87 (6) Å3 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 977 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed x-ray tube | Rint = 0.027 |
| Graphite monochromator | θmax = 27.5°, θmin = 3.1° |
| Detector resolution: 10 pixels mm-1 | h = −6→5 |
| ω scans | k = −10→10 |
| 3738 measured reflections | l = −12→12 |
| 1725 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.054 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.234 | H-atom parameters constrained |
| S = 1.16 | w = 1/[σ2(Fo2) + (0.1038P)2 + 0.301P] where P = (Fo2 + 2Fc2)/3 |
| 1725 reflections | (Δ/σ)max < 0.001 |
| 111 parameters | Δρmax = 0.39 e Å−3 |
| 0 restraints | Δρmin = −0.44 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3162 (6) | 0.2759 (3) | 0.1180 (3) | 0.0304 (7) | |
| H1 | 0.2882 | 0.1685 | 0.082 | 0.036* | |
| C2 | 0.5104 (6) | 0.3236 (3) | 0.2269 (3) | 0.0282 (7) | |
| C3 | 0.5524 (6) | 0.4852 (3) | 0.2811 (3) | 0.0279 (6) | |
| C4 | 0.3981 (6) | 0.5939 (3) | 0.2252 (3) | 0.0293 (7) | |
| H4 | 0.425 | 0.7012 | 0.2616 | 0.035* | |
| C5 | 0.2021 (6) | 0.5460 (3) | 0.1150 (3) | 0.0264 (6) | |
| C6 | 0.1606 (6) | 0.3872 (3) | 0.0611 (3) | 0.0274 (6) | |
| C7 | −0.0418 (6) | 0.3343 (3) | −0.0564 (3) | 0.0286 (6) | |
| O1 | −0.0753 (5) | 0.1938 (3) | −0.1056 (2) | 0.0421 (6) | |
| C8 | 0.6538 (9) | 0.0647 (4) | 0.2327 (4) | 0.0529 (10) | |
| H8A | 0.4579 | 0.0121 | 0.2389 | 0.079* | |
| H8B | 0.7865 | 0.0095 | 0.2828 | 0.079* | |
| H8C | 0.7052 | 0.0628 | 0.1374 | 0.079* | |
| O2 | 0.6714 (5) | 0.2264 (2) | 0.2899 (2) | 0.0369 (6) | |
| C9 | 0.8062 (7) | 0.6813 (4) | 0.4442 (3) | 0.0382 (8) | |
| H9A | 0.8839 | 0.7476 | 0.3745 | 0.057* | |
| H9B | 0.946 | 0.6892 | 0.5202 | 0.057* | |
| H9C | 0.6283 | 0.7173 | 0.4764 | 0.057* | |
| O3 | 0.7479 (5) | 0.5185 (2) | 0.3876 (2) | 0.0345 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0295 (15) | 0.0273 (15) | 0.0345 (15) | 0.0050 (11) | −0.0052 (12) | 0.0030 (12) |
| C2 | 0.0270 (14) | 0.0262 (14) | 0.0325 (14) | 0.0079 (11) | −0.0031 (12) | 0.0033 (11) |
| C3 | 0.0277 (14) | 0.0278 (15) | 0.0280 (14) | 0.0026 (11) | −0.0024 (11) | 0.0040 (11) |
| C4 | 0.0292 (14) | 0.0235 (14) | 0.0348 (15) | 0.0034 (11) | −0.0048 (12) | 0.0022 (11) |
| C5 | 0.0254 (14) | 0.0226 (14) | 0.0320 (14) | 0.0048 (11) | 0.0015 (12) | 0.0039 (11) |
| C6 | 0.0274 (14) | 0.0254 (15) | 0.0299 (14) | 0.0045 (11) | 0.0015 (12) | 0.0037 (11) |
| C7 | 0.0262 (14) | 0.0255 (14) | 0.0342 (15) | 0.0040 (11) | −0.0034 (12) | 0.0030 (11) |
| O1 | 0.0474 (14) | 0.0269 (12) | 0.0514 (14) | 0.0090 (9) | −0.0156 (11) | −0.0022 (9) |
| C8 | 0.062 (2) | 0.0276 (17) | 0.070 (2) | 0.0148 (15) | −0.025 (2) | −0.0001 (16) |
| O2 | 0.0405 (12) | 0.0287 (12) | 0.0423 (12) | 0.0116 (9) | −0.0126 (10) | 0.0013 (9) |
| C9 | 0.0418 (18) | 0.0317 (17) | 0.0392 (16) | 0.0025 (13) | −0.0133 (14) | −0.0007 (13) |
| O3 | 0.0377 (12) | 0.0289 (11) | 0.0356 (11) | 0.0049 (8) | −0.0145 (9) | −0.0002 (8) |
Geometric parameters (Å, º)
| C1—C2 | 1.382 (4) | C6—C7 | 1.474 (4) |
| C1—C6 | 1.404 (4) | C7—O1 | 1.236 (3) |
| C1—H1 | 0.94 | C7—C5i | 1.479 (4) |
| C2—O2 | 1.359 (3) | C8—O2 | 1.428 (4) |
| C2—C3 | 1.414 (4) | C8—H8A | 0.97 |
| C3—O3 | 1.356 (3) | C8—H8B | 0.97 |
| C3—C4 | 1.379 (4) | C8—H8C | 0.97 |
| C4—C5 | 1.397 (4) | C9—O3 | 1.434 (3) |
| C4—H4 | 0.94 | C9—H9A | 0.97 |
| C5—C6 | 1.392 (4) | C9—H9B | 0.97 |
| C5—C7i | 1.479 (4) | C9—H9C | 0.97 |
| C2—C1—C6 | 120.2 (3) | O1—C7—C6 | 121.0 (2) |
| C2—C1—H1 | 119.9 | O1—C7—C5i | 120.7 (2) |
| C6—C1—H1 | 119.9 | C6—C7—C5i | 118.3 (2) |
| O2—C2—C1 | 125.2 (3) | O2—C8—H8A | 109.5 |
| O2—C2—C3 | 114.9 (2) | O2—C8—H8B | 109.5 |
| C1—C2—C3 | 119.9 (2) | H8A—C8—H8B | 109.5 |
| O3—C3—C4 | 125.5 (3) | O2—C8—H8C | 109.5 |
| O3—C3—C2 | 114.8 (2) | H8A—C8—H8C | 109.5 |
| C4—C3—C2 | 119.7 (2) | H8B—C8—H8C | 109.5 |
| C3—C4—C5 | 120.5 (3) | C2—O2—C8 | 117.2 (2) |
| C3—C4—H4 | 119.7 | O3—C9—H9A | 109.5 |
| C5—C4—H4 | 119.7 | O3—C9—H9B | 109.5 |
| C6—C5—C4 | 119.9 (3) | H9A—C9—H9B | 109.5 |
| C6—C5—C7i | 120.8 (2) | O3—C9—H9C | 109.5 |
| C4—C5—C7i | 119.3 (2) | H9A—C9—H9C | 109.5 |
| C5—C6—C1 | 119.7 (3) | H9B—C9—H9C | 109.5 |
| C5—C6—C7 | 120.9 (2) | C3—O3—C9 | 117.4 (2) |
| C1—C6—C7 | 119.3 (3) | ||
| C6—C1—C2—O2 | 179.8 (3) | C4—C5—C6—C7 | −179.1 (3) |
| C6—C1—C2—C3 | 0.0 (4) | C7i—C5—C6—C7 | 0.6 (5) |
| O2—C2—C3—O3 | 0.2 (4) | C2—C1—C6—C5 | −0.1 (4) |
| C1—C2—C3—O3 | −180.0 (3) | C2—C1—C6—C7 | 179.0 (3) |
| O2—C2—C3—C4 | −179.5 (3) | C5—C6—C7—O1 | 178.8 (3) |
| C1—C2—C3—C4 | 0.3 (4) | C1—C6—C7—O1 | −0.3 (4) |
| O3—C3—C4—C5 | 179.9 (3) | C5—C6—C7—C5i | −0.6 (5) |
| C2—C3—C4—C5 | −0.4 (4) | C1—C6—C7—C5i | −179.7 (3) |
| C3—C4—C5—C6 | 0.3 (4) | C1—C2—O2—C8 | 4.9 (5) |
| C3—C4—C5—C7i | −179.4 (3) | C3—C2—O2—C8 | −175.3 (3) |
| C4—C5—C6—C1 | 0.0 (4) | C4—C3—O3—C9 | −2.1 (4) |
| C7i—C5—C6—C1 | 179.7 (3) | C2—C3—O3—C9 | 178.2 (3) |
Symmetry code: (i) −x, −y+1, −z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C8—H8A···O1ii | 0.97 | 2.58 | 3.391 (5) | 142 |
| C9—H9B···O2iii | 0.97 | 2.54 | 3.494 (4) | 168 |
Symmetry codes: (ii) −x, −y, −z; (iii) −x+2, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GG2091).
References
- Boldt, P. (1967). Chem. Ber. 100, 1270–1280.
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Kitamura, C., Akamatsu, N., Yoneda, A. & Kawase, T. (2009). Acta Cryst. E65, o324. [DOI] [PMC free article] [PubMed]
- Ohta, A., Hattori, K., Kusumoto, Y., Kawase, T., Kobayashi, T., Naito, H. & Kitamura, C. (2012). Chem. Lett. 41, 674–676.
- Rigaku (1998). PROCESS-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku (1999). RAPID-AUTO Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812033119/gg2091sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812033119/gg2091Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812033119/gg2091Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




