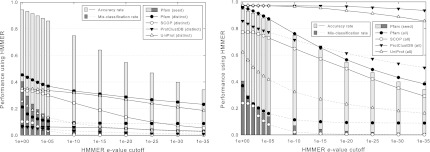
FIG. 4.
Accuracy rate and mis-classification rate of HMMER over different e-value cutoffs. Light bars and solid lines represent accuracy rates, while dark bars and dotted lines represent mis-classification rates. Bars denote performance on Pfam by applying HMMER on the profile hidden Markov model that was created for each curated family from a subset of curated seed sequences in Bateman et al. (2000). Lines denote performance by applying HMMER on the profile hidden Markov model that is constructed for each family in the training set from a multiple sequence alignment obtained by ClustalW. These alignments are constructed either from a subset of distinct sequences in each family with BLAST e-values above 0.1 (a BLAST e-value is considered to be above the cutoff if no hits are obtained) or from all sequences in each family. More tests are performed for e-value cutoffs between 1 and 1e–5.