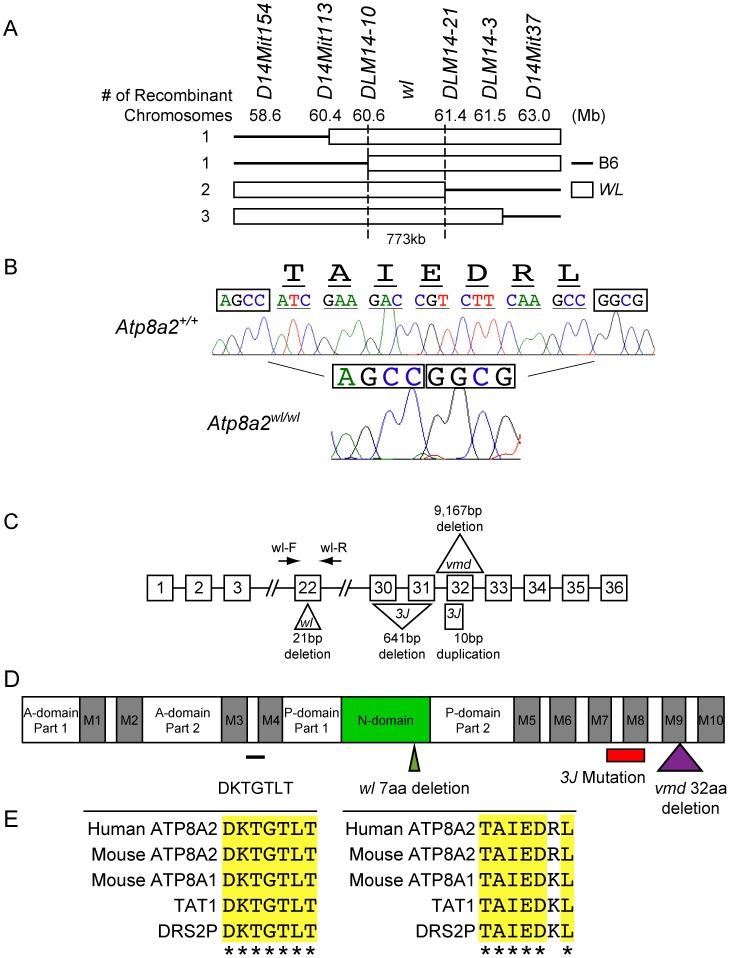
Figure 6. Positional cloning of the wl mutation.
(A) Genotyping 688 affected F2 mice (1354 meiotic events) allowed the wl locus to be mapped to a 773 kb region on mouse chromosome 14 between DLM14-10 (60.6 Mb) and DLM14-21 (61.4 Mb). This region contains 10 genes. Solid lines represent parts of the chromosome containing B6 sequences, while open rectangles symbolize parts of the chromosome containing wl sequences in different mice. Numbers on the left side refer to the number of independent recombinant chromosomes obtained. (B) Sequence analysis of genomic DNA from mutant (wl/wl) and wild type (+/+) mice revealed a 21 bp deletion (ATCGAAGACCGTCTTCAAGCC) in exon 22 of Atp8a2, while the rest of the coding sequence is still in frame. Nucleotide bases immediately flanking the deletion region are boxed for easy comparison. The amino acids (TAIEDRL) encoded by the deleted base pairs are given at the top of the sequences. (C) Diagram showing the position and nature of the wl mutation and two additional wl alleles, wlvmd (vmd) and wl3J (3J). vmd genomic DNA harbors a 9,167 bp deletion, resulting in loss of exon 32 from the RNA transcript (Figure S10). 3J has a 641 bp deletion starting from tenth base pair of exon 30. 3J also contains a duplication of 10 bp (TCTTTGGTGT) in exon 32 (Figure S11). (D) Protein domains of ATP8A2 with the location of the three wl allele mutations indicated. ATP8A2 contains 10 putative transmembrane domains (M1–M10), an actuator domain (A-domain), a nucleotide-binding domain (N-domain), and a phosphorylation domain (P-domain). The position of wl mutation resulting in the deletion of seven highly conserved amino acids (TAIEDRL) in the N-domain is indicated by a green triangle; the position of the vmd mutation resulting in the deletion of 32 amino acids spanning the ninth transmembrane domain is indicated by a purple triangle, while the area of the protein affected by the 3J deletion is indicated by a red bar (the small 3J duplication overlaps the vmd mutation). The location of seven highly conserved amino acids (DKTGTLT) is indicated with a black line. (E) Conservation of the seven amino acid DKTGTLT motif, which is highly conserved among all ATPases, as well as the seven amino acid sequence (TAIEDRL) deleted in wl, between mouse, human, C. elegans (TAT1) and yeast (DSR2P).