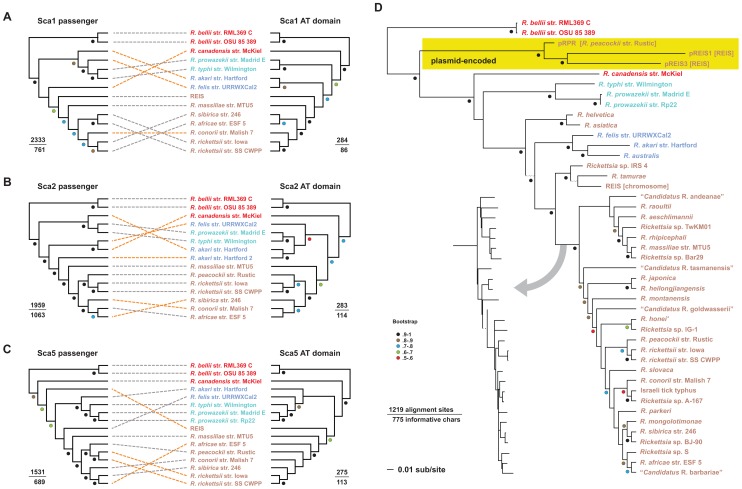
Figure 6. Evolution of rickettsial surface cell antigens (Scas).
All datasets were aligned with MUSCLE v3.6 [74], [75] and analyzed under maximum likelihood using RAxML [76] (see text for details). Branch support was assessed with 1000 bootstrap pseudoreplications, with values in intervals of 10 (>50% only) depicted with colored circles that are explained at the bottom left. Taxon color scheme according to major rickettsia group is explained in the Figure 2 legend. (A–C) Comparison of the phylogenies estimated for the passenger (left) and AT domains (right) for (A) Sca1, (B) Sca2 and (C) Sca5. For each tree, numbers above the line depict the total number of positions for each domain alignment, with numbers under the line representing the total number of informative sites within the alignment. The phylogenies of the AT domain better corroborate the rickettsia species tree (see Figure 2), thus the position of the taxa correspond with these trees. The taxa in the trees generated from the passenger domain are connected to their corresponding partner in the AT domain trees via dashed lines, with orange illustrating the major differences between the domain-generated trees. (D) Phylogeny estimation of an expanded set of rickettsial Sca4 proteins. The yellow box encloses the only known plasmid-encoded Sca4 proteins: pREIS1 and pREIS3 from REIS, rRPR from R. peacockii [83]. The gray arrow illustrates the replacement of the phylogram portion of the tree with the corresponding cladogram to account for the minimal divergence across the majority of SFG Rickettsia Sca4 proteins.