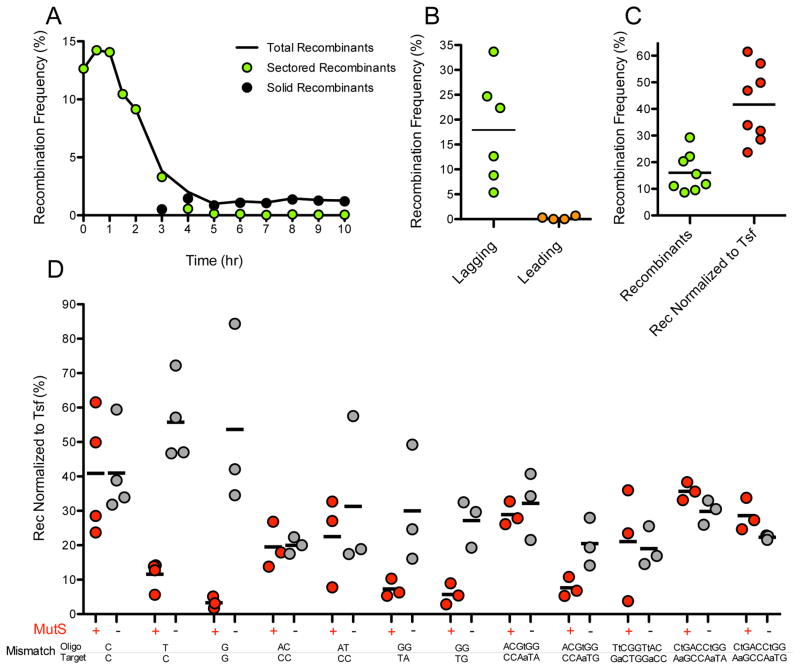
Figure 2. ssDNA Recombineering can be very efficient.
Unless specified, strain RIK423 was recombined with the lagging strand 99-base oligo 11. Recombination frequency is reported in each panel as the % of green colonies among the total number of colonies. (A) Segregation of recombinants during outgrowth. Sectored recombinants are colonies with a mixture of recombinant and non-recombinant cells. Solid recombinants are colonies composed of recombinant cells only. (B) Strand bias in targeting the lagging or leading strand template. Recombination targeting the lagging strand template (oligo 11 × RIK423) was 72 times more efficient than recombination targeting the leading strand template (oligo10 × RIK410). (C) The average recombineering frequency approaches 40% when normalized to transformation frequency. (D) MMR can be a potent inhibitor of recombination. MMR was evaluated in isogenic MMR+ and MMR− (ΔmutS::Kan) strains. A one-tailed unpaired t test with Welch’s correction was performed for each of the MutS+ and MutS− pairs. Additional details are given in the Supporting Information.