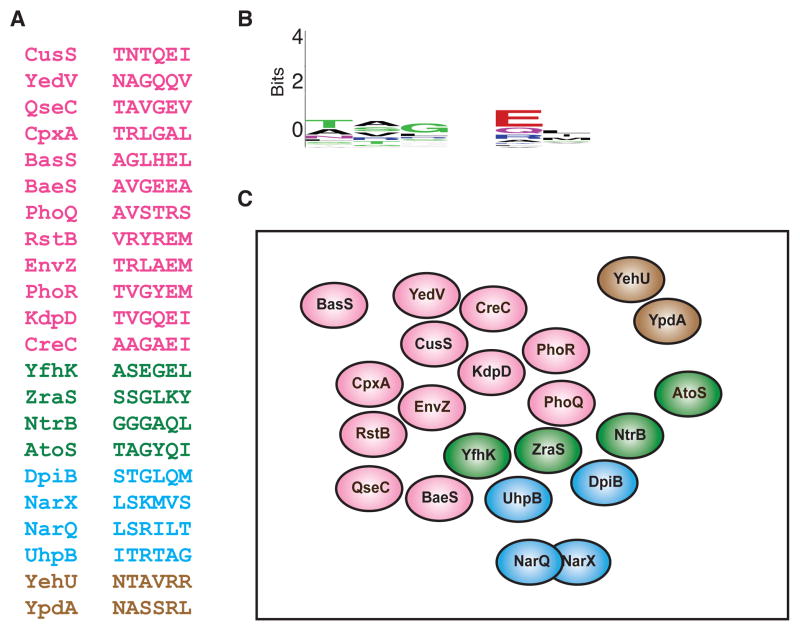
Figure 5. Extant two-component signaling pathways are insulated from each other at the level of phosphotransfer.
(A) The six primary specificity residues are shown for each of the 22 canonical E. coli histidine kinases. Hybrid histidine kinases and the non-canonical kinases DcuS and CheA were omitted. The histidine kinases are separated into groups by color based on the family of their cognate response regulator: pink, OmpR/winged helix-turn helix; green, NtrC/AAA+ and Fis domains; blue, NarL/GerE helix-turn-helix; brown, LytR. For specificity residues from E. coli response regulators and C. crescentus histidine kinases and response regulators, see Figure S4. (B) Sequence logo for the specificity residues in panel A. (C) A qualitative two-dimensional representation of the distribution of E. coli histidine kinases in the sequence space defined by the six primary specificity-determining residues. Each oval represents the set of response regulators recognized by a histidine kinase given its specificity residues. Spheres are colored using the same scheme as in panel A. With the exception of NarQ and NarX (see text), the spheres are non-overlapping, indicating a lack of cross-talk in vivo and in vitro. Kinases were placed relative to one another based roughly on their ability to phosphorylate the cognate regulators of other histidine kinases after extended incubation times in vitro (Skerker et al., 2005; Yamamoto et al., 2005). For example, CpxA shows a strong preference for phosphotransfer to its cognate regulator CpxR, but will phosphorylate the cognate regulators of EnvZ and RstB after extended periods of time.