Abstract
SMC (structural maintenance of chromosomes) complexes function ubiquitously in organizing and maintaining chromosomes. Functional fluorescent derivatives of the Escherichia coli SMC complex, MukBEF, form foci that associate with the replication origin region (ori). MukBEF impairment results in mispositioning of ori and other loci in steady-state cells. These observations led to an earlier proposal that MukBEF positions new replicated sister oris. We show here that MukBEF generates and maintains the cellular positioning of chromosome loci independently of DNA replication. Rapid impairment of MukBEF function by depleting a Muk component in the absence of DNA replication leads to loss of MukBEF foci as well as mispositioning of ori and other loci, while rapid Muk synthesis leads to rapid MukBEF focus formation but slow restoration of normal chromosomal locus positioning.
INTRODUCTION
The bacterial chromosome is not only highly compacted but also spatially organized, with chromosomal regions occupying specific positions in the cell (26, 35). For example, in newly formed Escherichia coli cells, the replication origin region (ori) is located at midcell and the left and right replication arms (replichores) occupy opposite cell halves (left-ori-right), with replication regenerating this arrangement in sisters (23, 42). The E. coli SMC (structural maintenance of chromosomes) complex, MukBEF, appears to play an important role in the left-ori-right arrangement about the cell transverse axis, because this organization is lost in Muk− cells, with sister oris moving to the outer nucleoid edges and the replication termination regions (ter) being located close to midcell (6, 43). For brevity, we refer to this positioning in the cell as nucleoid organization.
This arrangement bears similarities to those in Caulobacter crescentus and Bacillus subtilis, where characterized processes can position oris at cell poles (2, 38, 46); in Vibrio cholerae, the primary chromosome also has a pole-proximal ori, directed by an uncharacterized mechanism (8).
SMC protein complexes share a distinctive conserved architecture and play a range of roles in chromosome maintenance and processing in all three domains of life (22, 45). Most bacteria have a single SMC complex comprising a homodimer of SMC interacting with two accessory subunits, ScpA (kleisin) and ScpB (9). E. coli and some other gammaproteobacteria use a distant relative of SMC, MukB, which acts with two non-SMC subunits, MukE and MukF (kleisin) (24, 47). Bacterial smc null mutants are frequently temperature sensitive, produce a significant fraction of anucleate cells, and are hypersensitive to gyrase inhibitors (6, 14, 29), suggesting that SMC complexes play roles in chromosome compaction and segregation. Consistent with this, fluorescence in situ hybridization (FISH) and fluorescence repressor-operator system (FROS) studies have shown that chromosomal regions are mispositioned in B. subtilis and C. crescentus as well as in E. coli in the absence of the SMC complex (6, 10, 17, 33).
In vivo, fluorescent derivatives of both MukBEF and SMC-ScpAB form foci that associate with ori (6, 10, 21, 33). In B. subtilis and Streptococcus pneumoniae, ori association is mediated by interaction of SMC with ParB and its association with parS sites distributed throughout ori (10, 21, 33). The molecular basis of the MukBEF association with the ∼400 kb ori is not known, although it is not obviously restricted to a small defined region (6). Furthermore, the mechanism of action of MukBEF on DNA is not understood, and it is unclear whether its role is confined to a particular stage of the cell cycle and whether it associates with regions of the chromosome in addition to ori. A number of studies have suggested roles for SMC/MukBEF in ensuring effective chromosome segregation and/or organization of newly replicated DNA (6, 10, 30, 33). Further evidence has come from a genetic interaction of SMC and topoisomerase IV (TopoIV) in B. subtilis (34) and from in vitro studies demonstrating a physical interaction between MukB and TopoIV (13, 19), suggesting that MukBEF could promote effective sister chromosome unlinking during segregation (40).
In this study, we set out to test if the action of MukBEF is restricted to a specific stage of a cell's lifetime. We show that rapid depletion of functional MukBEF leads to correlated changes in positioning of ori and distant loci in cells containing a single unreplicated chromosome and in replicating cells, independently of cell generation stage. Functional MukBEF repletion, obtained by induced expression of MukF in otherwise Muk− cells, leads to immediate formation of MukBEF foci and a slow repositioning of chromosomal loci. Thus, MukBEF, independently of DNA replication, is required to generate and maintain wild-type chromosome positioning in the cell, defined by the left-ori-right arrangement about the cell transverse axis.
MATERIALS AND METHODS
Bacterial strains and growth.
Strains and plasmids used in this study are listed in Table 1. All strains were derivatives of E. coli K-12 AB1157 (1). Chromosomal loci were visualized by the fluorescence repressor-operator system (41, 42). tetO arrays (240 copies) inserted at positions 18 kb counterclockwise (CCW) of oriC (ori1), 50 kb clockwise (CW) of dif (ter3), and L3 (2268kb) and lacO arrays (240 copies) at positions L2 (366 kb) and R3 (852 kb) were used. LacI-mCherry and TetR-mCerulean were expressed either chromosomally (27) or from a plasmid (42). In the case of plasmid expression, AT (anhydrotetracycline) (40 ng ml−1) and IPTG (isopropyl-β-d-thiogalactopyranoside; 0.5 mM) was added to reduce operator occupancy and avoid replication blockage (41, 42). Western blotting for MukE depletion was done on cells growing in LB using a polyclonal antibody to c-Myc (Sigma-Aldrich). For MukF, this was done using antibody to MukF (48).
Table 1.
Strains and plasmids
| Strain | Description or referencea |
|---|---|
| Ab19 | mukE degron; Kanr ΔsspB |
| Ab62 | mukE degron FRT; ΔsspB Para sspB lacO at ori1 Kanr tetO at lacZ Gmr (LacI-CFP and TetR-YFP expressed from pWX6) |
| Ab64 | mukE degron FRT; ΔsspB Para sspB lacO at ori1 Kanr dnaC2(Ts) Tetr (LacI-mCherry expressed from pWX17) |
| Ab82 | mukE degron FRT; ΔsspB Para sspB lacO at L3 Kanr tetO at R3 Gmr dnaC2(Ts) Tetr (LacI-CFP and TetR-YFP expressed from pWX6) |
| Ab86 | mukE degron FRT; mukB-mYPet Kanr ΔsspB Para sspB tetO at ori1 Gmr (TetR-mCerulean expressed from pWX9) |
| Ab174 | ΔmukF::Kan mukB-gfp Cmr Para mukF FRT tetO at ori1 Gmr PlacI tetR FRT dnaC2(Ts) Tetr |
| Ab188 | ΔmukF::Kan tetO at ori1 Gmr PlacI tetR FRT dnaC2(Ts) Tetr |
| Ab229 | ΔmukF::Kan Para mukF FRT lacO at L3 Kanr tetO at R3 dnaA46(Ts) Tetr (LacI-CFP and TetR-YFP expressed from pWX6) |
| Ab234 | ΔmukF::Kan mukB-gfp Cmr Para mukF FRT lacO at ori1 tetO at ter3 |
| RRL48 | tetO at ori1 Gmr PlacI tetR FRT |
| pAb41 | pBAD24+sspB |
| pWX6 | 41 |
| pWX9 | 41 |
| pWX17 | 41 |
FRT, Flp recombination target; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein. For details of array positions on the chromosome, see Materials and Methods.
Construction of the MukE degron.
Degron-tagged MukE was constructed in the chromosome at the original location of the gene by the process of λ-Red recombination (7). mukE was thus fused to a single c-Myc tag followed by the DAS+4-degron tag (20). The plasmid carrying a 6-amino-acid (aa) linker, the Myc tag, a 2-aa linker, and the DAS+4-degron tag (SAGSAAEQKLISEEDLSSAANDENYSENYADAS) was described previously (28). λ-Red recombination was carried out in an AB1157 strain with sspB deleted and carrying the plasmid pKD46. After fusion of the degron tag to the C terminus of mukE, the fusion was transduced into a strain with sspB deleted at the endogenous gene locus, but with an arabinose-inducible copy at an ectopic locus on the chromosome (20).
For overexpression of sspB from a plasmid, the gene was cloned into pBAD24 between the NheI and KpnI sites. Degron constructs were checked for loss of viability at 37°C by streaking on plates with 0.2% arabinose. Plasmid expression of SspB was repressed by the addition of 0.2% glucose.
MukB fused to m-YPet or green fluorescent protein (GFP) at its C terminus was also constructed at the endogenous chromosome position by λ-Red recombination following the strategy described in reference 27. By all criteria this fusion was fully functional, and the foci have physiological significance (data not shown).
Construction of Para-mukF.
The strain carrying the MukF repletion system has been described previously (31). An ectopic copy of mukF FRT under the control of the Para promoter was introduced at the argE locus in AB1157. This was then transduced into an AB1157 strain carrying a deletion of mukF (replaced by kanamycin) in the operon (47).
Microscopy.
Cells were visualized with a 100× objective on a Nikon Eclipse TE2000-U microscope, equipped with a Photometrics Cool-SNAP HQ charge-coupled device (CCD) camera. Images were taken and processed by Metamorph 6.2. Image analysis was done using ImageJ or MicrobeTracker (32) run in Matlab.
Cells were grown at 30°C or 37°C in M9 medium supplemented with 0.2% glycerol (M9-gly) and were shifted to 37°C or 42°C (in the case of dnaC(Ts) or dnaA(Ts) mutants, respectively) to prevent new rounds of replication initiation. Δmuk cells were grown at 22°C (generation time in M9-gly was ∼4 h). Induction of expression from an arabinose-inducible promoter (11, 18) was done by the addition of 0.2 to 0.5% arabinose. For microscopy, cells in exponential phase (A600 0.05 to 0.2) were concentrated and laid on a 1% agarose pad on a slide. Nucleoids were visualized using a final concentration of 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Cell division was blocked by the addition of cephalexin (100 μg/ml).
MukE degradation experiment in replication-blocked cells.
Degron-mediated MukE degradation used a chromosomally expressed Para sspB. Cells were grown to an A600 of 0.05 in M9-gly at 30°C to ensure nonoverlapping replication cycles; the mass doubling times were 130 min (41). Cells were shifted to 37°C for 2 h, where the doubling time was ∼100 min. At the end of 2 h, a control sample (0 min) was collected and 0.2% arabinose was added to the culture at 37°C. Samples were collected 2 h after arabinose induction. All samples were spun down immediately at 37°C, spotted on a prewarmed slide, and imaged at 37°C.
MukF repletion experiment in replication-blocked cells.
Cells were grown in M9-gly at 30°C overnight. A dilution of the overnight culture was made into M9-gly and grown till A600 of 0.05. Cells were shifted to 37°C [for dnaC2(Ts)] or 42°C [for dnaA46(Ts)] for 2 h. A sample of cells (0 min) was taken before the addition of 0.5% arabinose to the culture. Samples were also taken 20, 40, 60, and 120 min after arabinose addition. All samples were spun down at 37°C or 42°C, spotted on a prewarmed slide and imaged at a restrictive temperature under the microscope. For time-lapse experiments, the same experimental setup was followed, but no arabinose was added to the culture; rather, cells were spotted on a slide containing medium with 0.5% arabinose. All imaging was done at 37°C.
MukF repletion in steady-state cells.
Cells were grown in M9-gly at 30°C overnight. A dilution of the overnight culture was made in M9-gly and grown until an A600 of 0.05 was reached. A sample of cells (0 min) taken before the addition of 0.5% arabinose to the culture. Samples were also taken 1, 10, 20, 40, 60, and 120 min after arabinose addition. The 1-min time point is effectively 2 to 3 min later (including the time taken for sampling and for imaging). All samples were spun down, spotted on a prewarmed slide, and imaged under the microscope. For time-lapse experiments, the same experimental setup was followed, but no arabinose was added to the culture; rather, cells were spotted onto a slide that contained medium with 0.5% arabinose.
Image analysis. (i) Assessing the position of chromosomal loci.
In order to estimate the positions of chromosomal loci, the Point Picker plugin (http://bigwww.epfl.ch/thevenaz/pointpicker/) of ImageJ was used. In cells with two copies of each locus, the x,y coordinates for the two cell poles and for the two loci were marked. From these, the absolute distances (in nm) of two loci from each cell pole as well as the relative distances, expressed as percentage of cell length, were estimated. For cells with two copies of each locus, as a rule, the cell pole farthest to the two loci was marked as the first pole (0%) and the opposite pole was marked as the end of the cell (100%). The average position and standard deviation (expressed as percentage of cell length) were plotted for each locus. Sister locus distance (SLD) was expressed as the average distance (percentage of cell length) between two sister loci in the same cell.
In cells with a single copy of each locus, the position of the locus was marked with respect to nucleoid pole. The nucleoid was divided into three categories: nucleoid middle (40 to 60% of nucleoid length), nucleoid quarter (60 to 80% of nucleoid length), and nucleoid edge (80 to 100% of nucleoid length). As a rule, the pole farthest from the locus was marked as the first pole (0%) and the nearest pole as the end of the nucleoid (100%). Hence, no loci were present in the 0-to-40% half of the nucleoid. Interlocus distance was calculated similar to sister chromatid distance (expressed as percentage of nucleoid length).
(ii) Defining a MukB focus.
MukB foci were counted automatically using the Microbetracker's spotfinderZ program (32). In the case of MukF repletion experiments in steady-state cells (see Fig. S3D in the supplemental material), when MukB foci were very weak, estimation of foci was done manually by using line profiles of fluorescence distribution. For each cell, maximum intensity in the profile was normalized to 100%. A peak in fluorescence intensity >40% above background fluorescence and with a full width at half maximum not greater than 4 pixels was considered a focus (see Fig. S4 in the supplemental material).
(iii) Defining colocalization between a MukB focus and chromosomal locus.
Distance between MukB foci and chromosomal loci were calculated automatically using Microbetracker's Colocalize program (32). A MukB focus was considered to colocalize with a chromosomal locus if the distance between them was ≤250 nm (which is close to the resolution limit of our system).
RESULTS
MukE depletion leads to loss of normal chromosome locus positioning in the absence of DNA replication.
We previously showed that functional MukBEF is essential for the normal positioning of newly replicated oris and for maintaining the wild-type arrangement of E. coli chromosome arms (6). It was nevertheless unclear whether MukBEF generates and/or maintains this organization. Furthermore, it was unknown when during a given cell generation MukBEF acts and if this action is directed exclusively to newly replicated DNA. To address these questions, we developed systems to rapidly produce or inactivate one of the MukBEF components through controlled expression or degradation, respectively.
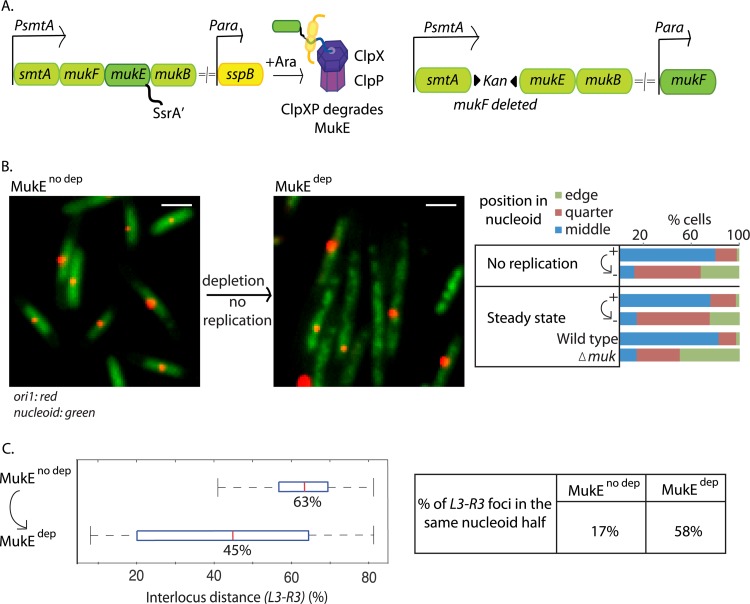
To test whether changes in locus positioning are dependent on ongoing DNA replication, functional MukBEF was impaired by degron-mediated depletion of MukE in cells containing a single nonreplicating chromosome (Fig. 1A). This was achieved by using dnaC(Ts) cells at 37°C, conditions that do not allow replication initiation, as judged by lack of replisomes, a single unsegregated ori, and flow cytometry, but do support completion of rounds of replication during most generations (40, 44). Cellular positions of ori1, L3, and R3 loci (42) (18 kb CCW, 1000 kb CCW, and 1000 kb CW of oriC, respectively) were analyzed before and 2 h after MukE depletion in nonreplicating cells. As expected, >85% cells contained a single ori1 focus as a consequence of their being unable to initiate replication from their single unreplicated chromosome (Fig. 1B). After 2 h of depletion (about one generation), ∼70% of the MukE had been degraded in steady-state cells, with most having lost their MukBEF foci, consistent with cells being MukBEF− phenocopies; indeed Muk− phenotypes were evident after 20 min of depletion (see Fig. S1 in the supplemental material). Before depletion, 80% of nonreplicating cells had a single ori1 focus at midnucleoid (Fig. 1B; mean position, 54% of nucleoid length). In contrast, after 2 h of MukE depletion, only 12% cells had ori1 at midnucleoid, with the mean position of ori1 having been shifted to 74% of nucleoid length. Along with the changes in ori1 positioning, the average L3-R3 interlocus distance (ILD) decreased from 63% to 45% after depletion, with 58% of L3 and R3 foci now in the same nucleoid half, rather than the 17% prior to depletion (Fig. 1C). Analysis of single ori1 focus steady-state cells also showed a clear shift of ori1 position from midnucleoid after MukE depletion, a pattern similar to that in cells deleted for MukF (Fig. 1B). We noted that after MukE depletion, the nucleoids took on a somewhat “lumpy” extended appearance, as assessed by DAPI staining (Fig. 1B; nucleoid length increased by ∼50%). We do not know if this is a direct or indirect consequence of MukBEF impairment, but it is not a direct consequence of induced SspB (data not shown). Indeed overexpression of SspB or presence of the SsrA tag on MukE did not affect nucleoid organization or cell growth (data not shown).
Fig 1.
MukE depletion leads to mispositioning of ori1 and L3-R3 independently of DNA replication. (A) Depletion and repletion. With depletion, arabinose-induced SspB directs MukE-SsrA′ to ClpXP for degradation (left). With repletion, MukF is expressed from an arabinose-inducible promoter at an ectopic chromosomal location in cells with wild-type MukF deleted (right). (B) MukE depletion results in ori mispositioning independently of replication. ori1 positions before (MukEno dep) and after (MukEdep) MukE depletion (2 h) in steady-state or nonreplicating [dnaC(Ts)] cells (strain Ab64) with a single ori1 focus are estimated as percentage of nucleoid length. Positions are binned into three categories: middle (40 to 60%), quarter (60 to 80%), or edge (80 to 100%) of nucleoid, and the percentage of cells with ori1 in each category is plotted. Wild-type and ΔmukF controls (strains RRL48 and Ab188, respectively) in steady-state cells with a single ori1 focus are also shown. Snapshot images of cells with labeled ori1 (red) and DAPI nucleoid staining (green) are shown before and after MukE depletion (in nonreplicating cells) on the left. n ≥ 300. Bars, 2 μm. (C) MukE depletion results in mispositioning of L3-R3 independently of replication. Interlocus distance (ILD; percentage of nucleoid length) between L3 and R3 before and after MukE depletion (2 h) in nonreplicating dnaC(Ts) cells (snapshot analysis) (strain Ab82). Red lines represent the median ILD. Boxes, interquartile ranges; whiskers, minima and maxima. Median values are shown below the red lines. The percentage of cells with L3 and R3 in the same nucleoid half is also shown (right). n ≥ 200.
We then examined segregated sisters of each locus by analyzing images of steady-state cells (see Fig. S2 in the supplemental material). The analysis showed reproducible changes after 2 h of MukE depletion. Sister locus distance (SLD) for sister ori1 loci increased, with sisters moving outwards from the normal quarter positions. In contrast, sisters of L3, R2, and R3, respectively, moved closer together after MukE depletion, almost always locating to the same cell half.
We conclude that impairment of MukBEF function by MukE depletion leads to changes in nucleoid organization that are independent of DNA replication. Therefore, MukBEF activity is required continuously throughout a cell's lifetime to maintain nucleoid organization.
Repletion of functional MukBEF leads gradually to normal locus positioning independently of DNA replication.
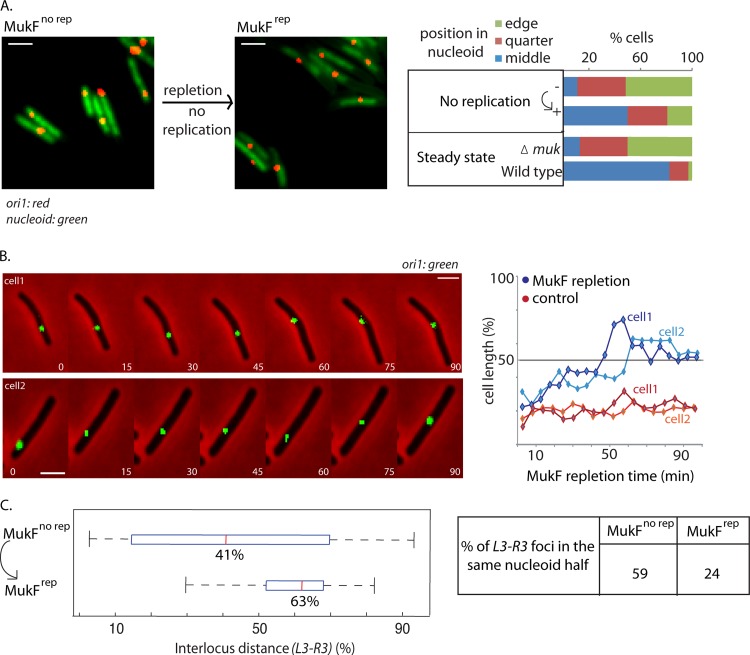
We also examined chromosome locus position when MukF expression was induced from an ectopic chromosomal locus in ΔmukF cells that had a single nonreplicating chromosome (Fig. 1A and 2A). Before induction of expression, 51% of cells had ori1 at the nucleoid edge and 11% of cells had ori1 at midnucleoid, with mean ori1 position at 79% of nucleoid length, comparable to the values for steady-state ΔmukF cells and for nonreplicating or steady-state MukE-depleted cells (Fig. 1B). After 1 h of induction, most cells that induced MukF expression now had ori1 at the wild-type midnucleoid position (Fig. 2A).
Fig 2.
MukF repletion repositions ori1 and L3-R3 in the absence of replication. (A) MukF repletion repositions ori1 independently of replication. ori1 position before (MukFno rep) and 1 h after (MukFrep) induction of MukF expression in nonreplicating dnaC(Ts) cells at 37°C (strain Ab174). Snapshot images of cells with labeled ori1 (red) and DAPI nucleoid staining (green) are shown before and after MukF repletion (in nonreplicating cells). n ≥ 300. Protocols, snapshot analysis, and controls were as described for Fig. 1. Bars, 2 μm. (B) ori1 repositioning during MukF repletion in the absence of replication. MukF repletion as a function of time [nonreplicating dnaC(Ts) cells]. Time-lapse analysis (strain Ab174) images were taken every 5 min for 100 min. The data for two representative cells are shown (blue lines), along with a subset of total images. Time-lapse traces of ori1 position for two representative nonreplicating ΔmukF cells are shown in red (strain Ab188). n ≥ 15. Bars, 2 μm. (C) MukF repletion repositions L3-R3 in the absence of replication. Interlocus distance (as percentage of nucleoid length) between L3 and R3 during MukF repletion (1 h) in nonreplicating [dnaA(Ts)] cells is shown (strain Ab229). The percentage of cells with L3 and R3 in the same nucleoid half is also shown (right). n ≥ 300.
Wild-type MukF protein levels were reached at 5 to 20 min of arabinose addition (see Fig. S1 in the supplemental material). The ∼20% of cells that retain ori1 close to the nucleoid edge had probably not induced efficient MukF expression, since we show later that a similar fraction of cells failed to respond to arabinose induction under the same conditions (Fig. 3A; also, see the supplemental material). Examination of ori1 position as a function of time of MukF expression in nonreplicating cells showed a gradual repositioning toward midnucleoid over the 60 min analyzed, although the induction of MukF and concomitant formation of MukBEF foci was rapid (Fig. 2A and B; also, see Fig. S1A and S3B, C, and D in the supplemental material). Before induction, only 11% cells had ori1 at midnucleoid; this had increased to 25% by 20 min of induction and to ∼50% after 1 h, by which time the nucleoids had returned to their wild-type length. Time-lapse analysis confirmed this MukF-dependent gradual repositioning of ori1 toward midnucleoid in nonreplicating cells (Fig. 2B; also, see Fig. S3C). Analysis of L3-R3 interlocus distances (ILD) also showed the complementary repositioning of L3-R3 during repletion (Fig. 2C). Prior to repletion, 59% of cells had L3 and R3 in the same nucleoid half, and mean L3-R3 ILD was 41%. After 1 h of repletion, mean L3-R3 ILD increased to 63%, with 24% of cells having L3 and R3 in the same nucleoid half. Similar slow repositioning of ori1 in steady-state cells containing a single ori1 focus was observed when either MukF or MukE expression was induced from an ectopic chromosomal locus in ΔmukF or ΔmukE cells, respectively (see Fig. S3A). We conclude that rapid expression of functional MukBEF in nonreplicating cells leads gradually, over ∼50% of a doubling time, to normal positioning of ori1 and other loci on both replichores.
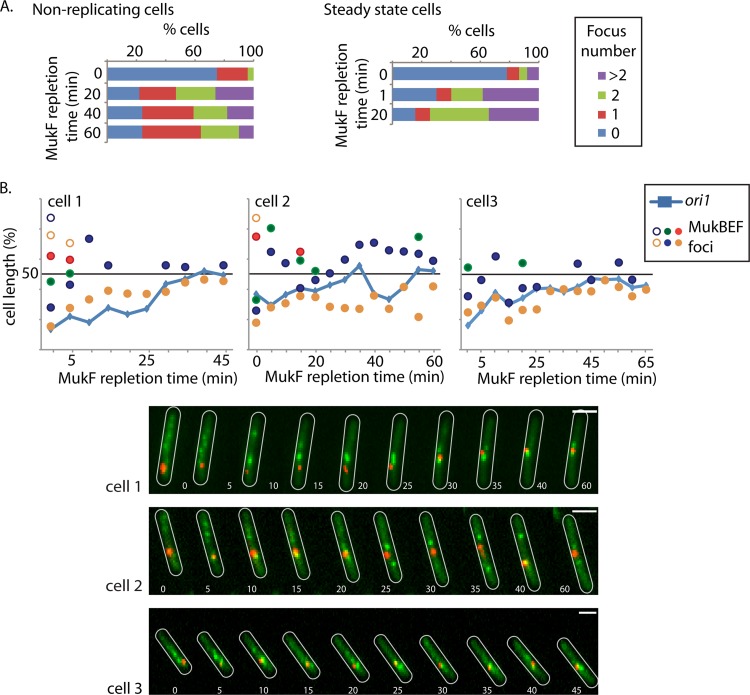
Fig 3.
Formation of MukB foci is independent of replication. (A) MukB focus appearance during MukF repletion is independent of replication or cell stage. The appearance and number of MukB foci were assessed before (0 min) and 20, 40, and 60 min after induction of MukF repletion (strain Ab174) in nonreplicating cells (left) and at 1 and 20 min after induction of MukF repletion in steady-state cells (strain Ab234). In the steady-state cells, transcription initiation was blocked at each time point by the addition of rifampin. The percentage of cells with 0, 1, 2, or >2 foci per cell are shown. n > 200. (B) MukB focus dynamics with respect to ori1 repositioning during MukF repletion in the absence of replication. Time-lapse analysis of MukB focus formation and ori1 positioning during MukF repletion in nonreplicating cells (strain Ab174). Time-lapse traces of ori1 (blue lines) as well as the position of MukB foci (circles) for three representative cells are shown. ori1, red; MukB, green. Cell outlines are in white. Images were taken every 5 min. Note that the 0-min time point in panel B is not equivalent to that in panel A, since there is a delay of ∼2 min between induction and imaging during the time-lapse experiments. Bars, 2 μm.
Formation of MukBEF foci is independent of replication.
MukBEF foci formed quickly after induction of MukF expression, in ΔmukF cells, irrespective of replication or cell size. Before induction, >75% of nonreplicating or steady-state cells had no clear MukBEF foci, and heterogeneous diffuse staining of MukB-GFP fluorescence was seen throughout the cell (Fig. 3A). Twenty-one percent of nonreplicating cells had at least one very weak MukBEF focus that may have arisen through low levels of arabinose-independent MukF expression (see Fig. S4 in the supplemental material for how a focus is defined). At 20 min after MukF induction, ∼75% of nonreplicating cells had MukBEF foci, with about a third of these having more than two foci. The situation was comparable after 20 min induction in steady-state cells (Fig. 3A). After 60 min of induction, ∼75% cells now had one or two MukBEF foci, as in steady-state wild-type cells (6), consistent with MukF overexpression not resulting in any aberrant behavior relative to MukBEF action. We infer that the ∼24% of cells that did not have MukBEF foci after 20, 40, or 60 min of incubation with arabinose had failed to induce efficient MukF expression.
Time-lapse analysis of MukBEF focus formation with respect to ori1 position in nonreplicating cells (Fig. 3B; also, see Fig. S3C in the supplemental material) confirmed that MukBEF foci appeared rapidly after induction of MukF expression and long before ori1 foci were normally repositioned close to midcell. Furthermore, several MukBEF foci distributed along the nucleoid were present at the earliest times, often with one being associated with ori1 during the repletion period, with the number returning to the wild-type level over time, consistent with the snapshot analysis. We do not know why this is, but we suspect that it is a consequence of re-establishing normal MukBEF function during repletion. Analysis of >300 randomly selected cells from an asynchronous population, which should represent all stages of a cell's life in a single generation, showed the appearance of foci in all cells within 5 min of MukF induction, indicating that MukBEF foci form independently of both replication and cell stage within a generation (see Fig. S3D in the supplemental material).
DISCUSSION
MukBEF directs a sequence of molecular events that shapes nucleoid organization.
We show here that the action of MukBEF in directing the normal positioning of ori and the two chromosome arms with respect to ori about the cell transverse axis is not restricted to newly replicated DNA. Furthermore, using degron-mediated MukE depletion, we also show that maintenance of the wild-type positioning of chromosomal regions is dependent on the continuous activity of MukBEF throughout the lifetime of a cell. Previously, we proposed that MukBEF directs the layering of newly replicated DNA on both sides of ori, resulting in the wild-type left-ori-right organization of the replichores with respect to ori about the cell transverse axis (6, 26). Similarly, others have proposed a close link between ongoing DNA replication and the action of MukBEF/SMC (29, 30). In contrast, the results presented here suggest that in E. coli there is no functional requirement for DNA replication for the action of MukBEF, with MukBEF foci forming independently of DNA replication and the stage within a cell's lifetime. In the absence of DNA replication, MukBEF-mediated repositioning of ori occurs slowly (≥30 min) in comparison with the few minutes it takes for MukBEF foci to form, and with the time required for the accumulation of normal levels of protein after induced MukF expression (5 to 20 min). Similarly, in steady-state cells, repletion of either MukF or MukE leads to a slow repositioning of ori.
We show that the formation of MukBEF foci, which are exclusively associated with the nucleoid (6), is not dependent on DNA replication. This contrasts with the cell cycle-regulated loading of cohesin and subsequent replication-dependent establishment of cohesion in eukaryotes (22). Although the B. subtilis and S. pneumoniae SMC complexes associate with ori through a ParB-parS interaction (10, 21, 33), the molecular basis of the ori association with MukBEF has not been identified.
Although the nature of the molecular transactions carried out by MukBEF remains unclear, we consider two non-mutually exclusive models, both of which assume that the MukBEF foci are important centers of MukBEF activity. In the first, MukBEF within foci plays a key role in positioning ori, consistent with MukBEF absence's leading to ori mispositioning (6). Furthermore, the action of MukBEF is restricted locally to ori, as observed through the ori1-MukBEF association in wild-type steady-state cells. The loss of the normal left-ori-right replichore organization would then be explained by correlated repositioning of other loci when the ori region moves, similarly to that reported in C. crescentus when the tethering point of the chromosome to the cell pole is moved (36). Such local action need not lead to any global changes to the chromosome other than through the left-ori-right replichore organization about the cell transverse axis. Since in this model MukBEF foci position oris, an ori-independent mechanism for the formation of MukBEF foci is required, for example, one similar to the mechanisms that can position plasmids and chemosensory machines at regular positions on the nucleoid (37). The early appearance of multiple MukBEF foci distributed throughout the nucleoid during repletion is consistent with locus-independent MukBEF positioning, as is the observation that MukBEF focus-ori association can be lost in other situations in which DNA metabolism, or MukBEF function, is perturbed (our unpublished data). Similarly, the slow correlated movement of ori1 with an associated MukBEF focus to the midcell position during repletion, along with a reduction in the number of MukBEF foci to the steady-state levels, underlines the importance of the ori-MukBEF association in correct ori positioning and indicates that restoration of the chromosome to a normal organization occurs slowly during functional MukBEF repletion. A corollary of this model is that correct ori positioning by MukBEF plays a role in faithful chromosome segregation.
The second hypothesis states that MukBEF has a direct global role in determining nucleoid structure and organization, with its association with ori helping direct organization of distant regions by a looping and/or bridging process. Loss of replichore organization in Muk− cells could then result from the breaking of MukBEF-mediated links between distant chromosomal regions. One could imagine this occurring by a “gathering in” of distant chromosomal regions by ori-associated MukBEF, similarly to the proposed action of H-NS, which interacts with multiple chromosome regions (39). The observation that at early times of repletion there are multiple MukBEF foci throughout the nucleoid (with one generally being ori associated), despite there being a single unreplicated copy of the chromosome, is also consistent with this hypothesis. MukBEF molecules in these foci could be in the gradual process of gathering in distant chromosome segments and associating them with ori. Others have proposed functions of SMC complexes in global compaction through bridging or SMC ring-mediated looping (4, 5, 12, 15, 25), while linkage between chromosomal regions has been proposed as a mechanism for the action of condensin in mitotic chromosomes (4). A global action on chromosomes has also been suggested for MukBEF and other SMCs through an effect on DNA topology, potentially facilitating compaction and segregation (3, 16, 29).
We now need a better understanding of the nature of the structures within MukBEF foci and how they form and associate with ori DNA. Insight from this work should help us understand how MukBEF and other SMC complexes function mechanistically in vivo.
Supplementary Material
ACKNOWLEDGMENTS
The work was funded by a Wellcome Trust Programme grant to D.J.S. R.R.-L. held the Todd-Bird Junior Research Fellowship of New College (Oxford).
We thank our many colleagues for discussion, in particular, Kim Nasmyth, Frank Uhlmann, and Steve Bell. We acknowledge Bing Li, who performed the first repletion experiments.
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 3. Carter SD, Sjogren C. 2012. The SMC complexes, DNA and chromosome topology: right or knot? Crit. Rev. Biochem. Mol. Biol. 47:1–16 [DOI] [PubMed] [Google Scholar]
- 4. Cuylen S, Haering CH. 2011. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 21:552–559 [DOI] [PubMed] [Google Scholar]
- 5. D'Ambrosio C, et al. 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 22:2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. 2007. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 65:1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fogel MA, Waldor MK. 2005. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol. Microbiol. 55:125–136 [DOI] [PubMed] [Google Scholar]
- 9. Gruber S. 2011. MukBEF on the march: taking over chromosome organization in bacteria? Mol. Microbiol. 81:855–859 [DOI] [PubMed] [Google Scholar]
- 10. Gruber S, Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137:685–696 [DOI] [PubMed] [Google Scholar]
- 11. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. 2008. The cohesin ring concatenates sister DNA molecules. Nature 454:297–301 [DOI] [PubMed] [Google Scholar]
- 13. Hayama R, Marians KJ. 2010. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:18826–18831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hiraga S, et al. 1991. Mutants defective in chromosome partitioning in E. coli. Res. Microbiol. 142:189–194 [DOI] [PubMed] [Google Scholar]
- 15. Hirano M, Hirano T. 2004. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 23:2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holmes VF, Cozzarelli NR. 2000. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. U. S. A. 97:1322–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jensen RB, Shapiro L. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. U. S. A. 96:10661–10666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241–3247 [DOI] [PubMed] [Google Scholar]
- 19. Li Y, et al. 2010. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc. Natl. Acad. Sci. U. S. A. 107:18832–18837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGinness KE, Baker TA, Sauer RT. 2006. Engineering controllable protein degradation. Mol. Cell 22:701–707 [DOI] [PubMed] [Google Scholar]
- 21. Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. 2011. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 81:676–688 [DOI] [PubMed] [Google Scholar]
- 22. Nasmyth K, Haering CH. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74:595–648 [DOI] [PubMed] [Google Scholar]
- 23. Nielsen HJ, Ottesen JR, Youngren B, Austin S, Hansen FG. 2006. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62:331–338 [DOI] [PubMed] [Google Scholar]
- 24. Niki H, et al. 1992. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 11:5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petrushenko ZM, Cui Y, She W, Rybenkov VV. 2010. Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. EMBO J. 29:1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reyes-Lamothe R, Wang X, Sherratt D. 2008. Escherichia coli and its chromosome. Trends Microbiol. 16:238–245 [DOI] [PubMed] [Google Scholar]
- 27. Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. 2008. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133:90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reyes-Lamothe R, Sherratt DJ, Leake MC. 2010. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328:498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sawitzke JA, Austin S. 2000. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl. Acad. Sci. U. S. A. 97:1671–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwartz MA, Shapiro L. 2011. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol. Microbiol. 82:1359–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sivanathan V, et al. 2009. KOPS-guided DNA translocation by FtsK safeguards Escherichia coli chromosome segregation. Mol. Microbiol. 71:1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80:612–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sullivan NL, Marquis KA, Rudner DZ. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137:697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tadesse S, Mascarenhas J, Kosters B, Hasilik A, Graumann PL. 2005. Genetic interaction of the SMC complex with topoisomerase IV in Bacillus subtilis. Microbiology 151:3729–3737 [DOI] [PubMed] [Google Scholar]
- 35. Toro E, Shapiro L. 2010. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2:a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Umbarger MA, et al. 2011. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell 44:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vecchiarelli AG, et al. 2010. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol. Microbiol. 78:78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viollier PH, et al. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:9257–9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang W, Li GW, Chen C, Xie XS, Zhuang X. 2011. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333:1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X, Reyes-Lamothe R, Sherratt DJ. 2008. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 22:2426–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X, Possoz C, Sherratt DJ. 2005. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 19:2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X, Liu X, Possoz C, Sherratt DJ. 2006. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20:1727–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weitao T, Dasgupta S, Nordstrom K. 2000. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol. Microbiol. 38:392–400 [DOI] [PubMed] [Google Scholar]
- 44. Withers HL, Bernander R. 1998. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 180:1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wood AJ, Severson AF, Meyer BJ. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11:391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu LJ, Errington J. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49:1463–1475 [DOI] [PubMed] [Google Scholar]
- 47. Yamanaka K, Ogura T, Niki H, Hiraga S. 1996. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250:241–251 [DOI] [PubMed] [Google Scholar]
- 48. Yamazoe M, et al. 1999. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18:5873–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.