Abstract
Cyanuric acid hydrolases (AtzD) and barbiturases are homologous, found almost exclusively in bacteria, and comprise a rare protein family with no discernible linkage to other protein families or an X-ray structural class. There has been confusion in the literature and in genome projects regarding the reaction products, the assignment of individual sequences as either cyanuric acid hydrolases or barbiturases, and spurious connection of this family to another protein family. The present study has addressed those issues. First, the published enzyme reaction products of cyanuric acid hydrolase are incorrectly identified as biuret and carbon dioxide. The current study employed 13C nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry to show that cyanuric acid hydrolase releases carboxybiuret, which spontaneously decarboxylates to biuret. This is significant because it revealed that homologous cyanuric acid hydrolases and barbiturases catalyze completely analogous reactions. Second, enzymes that had been annotated incorrectly in genome projects have been reassigned here by bioinformatics, gene cloning, and protein characterization studies. Third, the AtzD/barbiturase family has previously been suggested to consist of members of the amidohydrolase superfamily, a large class of metallohydrolases. Bioinformatics and the lack of bound metals both argue against a connection to the amidohydrolase superfamily. Lastly, steady-state kinetic measurements and observations of protein stability suggested that the AtzD/barbiturase family might be an undistinguished protein family that has undergone some resurgence with the recent introduction of industrial s-triazine compounds such as atrazine and melamine into the environment.
INTRODUCTION
With widespread DNA sequencing and complementary bioinformatics, more than 108 proteins are contained within the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/). The proteins, both known and virtual, have been assigned to family and superfamily compilations. Some of these compilations now include tens or hundreds of thousands of members. Many have multiple crystal structures available (2–4, 14, 28). Newly identified proteins typically map to one of these large superfamilies, thereby aiding in determining the new protein's structure and function.
The set of proteins identified as cyanuric acid hydrolases and barbiturases do not fit this typical pattern. The cyanuric acid hydrolases designated AtzD and TrzD were the first members of this family to be identified (12, 20). The atzD gene in Pseudomonas sp. strain ADP is part of an operon that encodes the catabolism of the s-triazine compound cyanuric acid (13, 26). Cyanuric acid catabolism releases ammonia to support cell growth. AtzD initiates catabolism by opening the s-triazine ring to allow subsequent ammonia-liberating reactions. TrzD is found in Acidovorax citrulli strain NRRLB-12227, a bacterium isolated for its ability to grow on melamine. TrzD is functionally identical to AtzD, catalyzing the cyanuric acid ring-opening reaction in the pathway. AtzD and TrzD were identified as being homologous, but they were not connected to any other proteins or protein families in those studies (12, 20, 26). Barbiturase was first purified to homogeneity from Rhodococcus erythropolis JCM3132 and shown to catalyze the hydrolytic opening of barbituric acid, an intermediate in the oxidative pyrimidine catabolic pathway (34, 35). Those studies reported that barbiturase was homologous to AtzD and TrzD (34, 35). One of the reports found zinc to be present in the enzyme preparations and thus proposed that barbiturase was homologous to members of the amidohydrolase superfamily (35). Barbiturase is not reactive with cyanuric acid. Conversely, cyanuric acid hydrolase does not hydrolyze barbituric acid, although that compound was observed to be a tight-binding inhibitor (11, 20).
Most recently, a few additional cyanuric acid hydrolases were experimentally confirmed in atrazine-degrading bacteria and from the genome sequence of Moorella thermoacetica ATCC 39073 (23). Additionally, several sequences have been annotated as cyanuric acid hydrolases or barbiturases from genome sequencing projects. Pairwise sequence comparisons of these new proteins typically show 40 to 65% protein sequence identity to AtzD, TrzD, or barbiturase, but all of these comparisons were similar. Thus, it was not possible to definitively discern whether a new sequence represented a cyanuric acid hydrolase, a barbiturase, or perhaps some other enzyme activity.
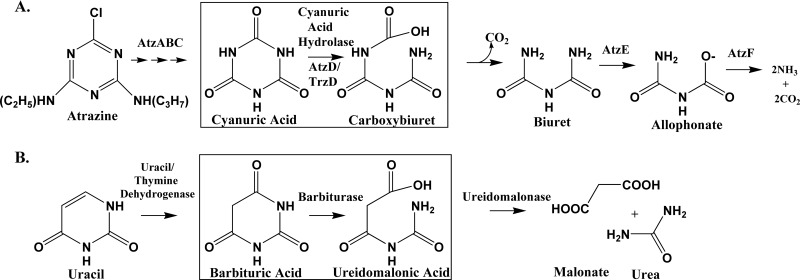
Cyanuric acid hydrolases and barbiturases, although clearly homologous, have been perceived as catalyzing somewhat different reactions. Barbiturase was reported to cleave a single amide bond in the six-membered ring of barbiturate to yield ureidomalonic acid (Fig. 1B). Purified cyanuric acid hydrolases have been reported by several groups (12, 20, 36) to yield biuret and carbon dioxide, which represents an apparent cleavage of two carbon to nitrogen bonds within the six-membered ring of cyanuric acid (Fig. 1A).
Fig 1.
Metabolic pathways showing the reactions catalyzed by cyanuric acid hydrolase (A) and barbiturase (B).
The present study dealt with these previous ambiguous features of the AtzD/barbiturase family and greatly expanded the set, with dozens of new related proteins being identified by bioinformatics. In total, 41 protein sequences were clustered into clades. Representative proteins from each clade were cloned, expressed, purified, and assayed to determine kinetic constants. This allowed assignment and, in a few cases, reassignment of proteins into clearly delineated classes as either cyanuric acid hydrolases or barbiturases. Moreover, the reactions catalyzed by cyanuric acid hydrolase and barbiturase were investigated and found to be directly analogous (Fig. 1). This correction of the reaction products is important because genomic and metabolism databases such as MetaCyc incorrectly describe the cyanuric acid hydrolase (AtzD and TrzD)-catalyzed reactions as amide hydrolysis and decarboxylation (http://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-5169) rather than the amide bond cleavage reaction alone, as shown here. Lastly, bioinformatics and metal determinations showed no evidence of homology between the AtzD/barbiturase family and the amidohydrolase superfamily.
MATERIALS AND METHODS
Chemicals.
Cyanuric acid, barbituric acid, 2,4,5-trihydroxypyrimidine, 6-azauracil, and hydantoin were purchased from Acros Organics (Morris Plains, NJ). Uracil, thymine, 5-azacytosine, cytosine, 5-nitrobarbituric acid, 5,6-dihydroxyuracil, allantoin, l-asparagine, biuret, and deuterium oxide (D2O) were purchased from Sigma-Aldrich (St. Louis, MO). Creatinine was purchased from Fluka (Buchs, Switzerland). Alloxan was purchased from Eastman Kodak (Rochester, NY). Urea and dioxane were purchased from Fisher Scientific (Pittsburgh, PA). Ammelide and ammeline were synthesized in our laboratory as previously described (31).
Identification of AtzD/barbiturase family members.
The nonredundant protein database was searched using pblast and PSI-BLAST with query sequences that included cyanuric acid hydrolase sequences from Pseudomonas sp. ADP (AtzD), Acidovorax citrulli strain 12227 (TrzD), and Moorella thermoacetica ATCC 39073 and the barbiturase sequence from Rhodococcus erythropolis JCM 3132. The data set was cross-referenced to ensure that all proteins were members of the same family. Shotgun analyses with both BLAST and PSI-BLAST were done in an attempt to broaden the sequence space of the AtzD/barbiturase family (30). The HMMER software (version 3.0) (9) was used to search the nonredundant protein database using Pfam hidden Markov models (HMMs), and HMMs were generated with the current data set.
Phylogenetic tree construction.
The sequences belonging to the AtzD/barbiturase family were collected and aligned using ClustalW (37). Phylogenetic trees were constructed using Phylip software (10). Maximum-likelihood and neighbor-joining methods were used with bootstrapping. The protein from Frankia sp. strain Eul1c had the most divergent sequence and was used as the outgroup. Changing outgroups did not alter the morphology of the trees.
The general clusterings found in maximum-likelihood and neighbor-joining trees were the same. Slight differences in position were observed for AtzD homologs from Bacillus cellulosilyticus DSM 2522, Paenibacillus sp. strain JDR-2, Acidithiobacillus ferrooxidans ATCC 53993 and ATCC 23270, Oceanicola granulosus HTCC2516, Saccharomonospora viridis DSM 43017, and Catenulispora acidiphila DSM 44928. These differences did not alter interpretation of the data.
Bacterial strains.
Strains were grown as recommended by the suppliers. Rhizobium leguminosarum bv. viciae 3841 was obtained from Peter Young (University of York, United Kingdom), Azorhizobium caulinodans ORS 571 was from Eric Giraud (Laboratoire des Symbioses Tropicales et Méditerranéennes, Montpellier, France), Moorella thermoacetica was from Stephen Ragsdale (University of Michigan Medical School, Ann Arbor, MI), Bradyrhizobium japonicum USDA 110 was from the USDA-ARS Rhizobium culture collection in Beltsville, MD, and DNA from Methylobacterium sp. strain 4-46 was from Chris Marx (Harvard University, Cambridge, MA) and Lynne Goodwin (DOE-Joint Genome Institute, Los Alamos National Laboratory).
Cloning, expression, and protein purification.
Genes encoding AtzD/barbiturase family members were PCR amplified and cloned into a pET28b+ vector (Novagen, Madison, WI), using NdeI and HindIII restriction sites. The resulting vectors were transformed into either Escherichia coli BL21(DE3) or E. coli BL21(DE3)pLys. These strains were grown with the appropriate antibiotics, i.e., 50 μg/ml kanamycin or 50 μg/ml kanamycin with 25 μg/ml chloramphenicol, respectively. Expression and protein purification were conducted as previously described (23). All enzymes that were purified were stored at 4°C and never frozen, with the exception of the Moorella thermoacetica ATCC 39073 enzyme used in nuclear magnetic resonance (NMR) experiments. Stability was monitored to ensure that no detectable inactivation occurred throughout the study.
Enzyme assays and kinetic constant determinations.
All reactions were carried out in 25 mM Tris-HCl buffer, pH 8.0, at room temperature unless otherwise noted. Assays were performed using a Beckman DU 640 spectrophotometer. Cyanuric acid and barbituric acid concentrations were monitored at 214 nm (extinction coefficient = 8,800 cm−1 M−1) and 256 nm, respectively. Substrates for routine assays were tested at 100 μM. Kinetic parameters were determined by obtaining rates at cyanuric acid concentrations ranging from 10 to 110 μM. Kinetic constants were determined by using the nonlinear regression analysis of Prism software (GraphPad, version 5.03).
The Azorhizobium caulinodans ORS 571 (locus AZC_3203) enzyme was tested with additional substrates, including uracil, thymine, 6-azauracil, 5-azacytosine, cytosine, 2,4,5-trihydroxypyrimidine, 5-nitrobarbaturic acid, creatinine, 5,6-dihydroxyuracil, hydantoin, ammelide, ammeline, allantoin, alloxan, urea, l-asparagine, and biuret. Transformation of these compounds consisted of noting any spectral changes via UV-visible spectrometry and any changes in peak areas via high-pressure liquid chromatography (HPLC) analysis, using a Hewlett-Packard HP 1100 system equipped with a diode array detector interfaced to an HP ChemStation. HPLC samples were prepared in 5 mM phosphate buffer, pH 7.0. Samples were analyzed on either an Alltech mixed-mode C8/anion 7 μ column (250 by 4.6 mm), using an isocratic mobile phase consisting of 95% methanol and 5% 5 mM phosphate buffer, pH 7.0; a Waters IC-Pak A HC anion-exchange column (150 by 4.6 mm), using an isocratic mobile phase consisting of 5 mM phosphate buffer, pH 8.0; or a Varian Microsorb-MV 100-8 CN column (250 by 4.6 mm), using an isocratic mobile phase of 5 mM phosphate buffer, pH 6.7.
Demonstration of cyanuric acid hydrolase reaction product via 13C NMR and mass spectrometry: detection of [U-13C]carboxybiuret.
Initially, broadband proton-decoupled 13C NMR spectra were acquired at 20°C and 100.5 MHz on a Varian Unity Inova 400 MHz NMR spectrometer equipped with a Nolorac 4 Nuc probe and operating under VnmrJ 2.2D software. 1,4-Dioxane was used as internal reference and integration standard (assigned as δ = 66.6 ppm relative to an external tetramethylsilane reference). Initial 13C NMR results were confirmed using a Varian Inova 600 MHz NMR spectrometer with a 5-mm Auto-X Dual Broadband probe held at 25°C.
Reactions were performed in 5-mm NMR tubes containing a 0.7-ml solution of uniformly ring-labeled [U-13C]cyanuric acid (2.0 mg/ml, 95% chemical purity, 99% isotopic purity; Toronto Research Chemicals, Inc., Ontario, Canada) in 100 mM potassium phosphate, 10% D2O, and 1% 1,4-dioxane (pH 8.9). The mixture had a final pH of ∼7.7. The 13C NMR spectrum of the [U-13C]cyanuric acid (singlet, 157.7 ppm) was stable overnight at room temperature. Spectra from commercial standards of biuret and bicarbonate were also acquired and had chemical shifts of 157.5 (singlet) and 160.4 ppm (singlet), respectively. Enzymatic reactions were initiated by adding 35 to 150 μg purified cyanuric acid hydrolase in 100 mM potassium phosphate buffer (pH 7.0) to the NMR tube. The cyanuric acid hydrolases used were from Pseudomonas sp. ADP, Moorella thermoacetica ATCC 39073, and Azorhizobium caulinodans ORS 571 (locus AZC_3892). Spectra representing the averaged chemical shifts from all pulses during the acquisition time were viewed at various intervals to follow the reaction process.
For mass spectrometry, 1-ml reaction mixtures contained 2.0 mg/ml cyanuric acid (unlabeled or U-13C labeled) in 100 mM ammonium acetate, pH 9.1. The pH with this mixture was 8.5. Reactions were initiated with the addition of 35 μl cyanuric acid hydrolase in 100 mM potassium phosphate, pH 7. Twenty-microliter samples were directly injected into a Waters high-resolution mass spectrometer in negative-ion mode at intervals over the course of 40 min with 3 ml/min water as the carrier fluid. Spectra were analyzed using the MassLynx software with Elemental Composition. Mass determinations were made in the following manner. Each ion between nominal masses of 100 and 500 was analyzed for its abundance over time. Those masses with profiles that reflected increasing abundance and then subsequent decreasing abundance were noted. The associated masses from the experiment with unlabeled cyanuric acid were then compared to those from the experiment with [U-13C]cyanuric acid to generate a list of paired ions, where the difference between their masses could be attributed to the difference between 12C and 13C. For each of the paired ions selected, the average mass from each time point was averaged and the standard deviation determined (Microsoft Excel 2003) to obtain the mass and tolerance to use in the Elemental Composition software.
Metal analysis.
The cyanuric acid hydrolases from Pseudomonas sp. ADP, Moorella thermoacetica ATCC 39073, and Bradyrhizobium japonicum USDA 110 and the barbiturase from Rhodococcus erythropolis JCM 3132 were hydrolyzed with 6 M hydrochloric acid at 110°C for 24 h under vacuum. After cooling, the samples were quantitatively transferred to a vial and diluted to 5 ml with Chelex-treated water. Metal concentrations were determined via inductively coupled plasma emission spectroscopy (ICP) at the Soil Analytical Laboratory of the University of Minnesota (St. Paul, MN). Protein concentrations were quantitatively ascertained by amino acid analysis at Purdue Proteomic Facility (West Lafayette, IN).
RESULTS
Identification of new members in the cyanuric acid hydrolase (AtzD)/barbiturase family.
Sequences from over 6,423 genomes (24) containing over 10 million genes were analyzed in an effort to expand the set of known cyanuric acid hydrolases/barbiturases and link them to other protein families using BLAST, PSI-BLAST, Shotgun, and HMMER. Efforts to link this family with another family or superfamily were unsuccessful. However, the effort did identify 41 homologous sequences as of 4 January 2011. Of those, 8 were identified in bacteria that metabolize s-triazine compounds, 32 were identified from genome sequencing data, and 1 was determined independently.
The cyanuric acid hydrolase/barbiturase proteins consist of 340 to 400 amino acids and have subunit molecular masses of 36 to 43 kDa. The sequence diversity was large, with most proteins 40 to 65% identical to any of the proteins of known function. The identified sequences were found in a diverse set of organisms that represented one eukaryotic (one member only) and five bacterial phyla. These organisms were found in a wide range of environments, which included soils, acid mine drainage, plant tissue, and human feces. Additionally, the source organisms had been studied for a diverse set of functions, including plant symbioses, photosynthesis, s-triazine degradation, and plant pathogenesis.
Phylogenetic analysis of protein sequences.
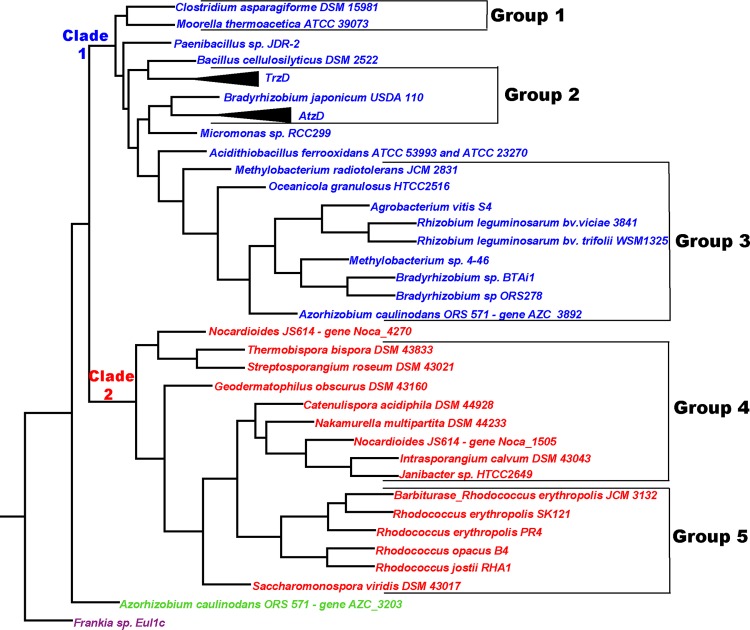
Phylogenetic analysis of the cyanuric acid hydrolase/barbiturase sequences generated a tree in which 39 of the 41 sequences were placed into two major clades (Fig. 2). Clade 1 contained 24 sequences, of which 20 were subdivided into 3 groups (groups 1, 2, and 3) based on branching patterns. Clade 2 contained 15 sequences, of which 14 were organized into 2 groups (groups 4 and 5) based on branching patterns and gene context (see Fig. S1 in the supplemental material). Sequences that varied their position depending on the tree-building algorithm used were left out of the defined groups. Clade 1 sequences were more divergent than clade 2 sequences. Most of the clade 1 sequences were only ∼40 to 65% identical to known functional family members, whereas all of the clade 2 sequences were ≥72% identical to the characterized barbiturase (Table 1).
Fig 2.
Maximum-likelihood phylogenetic tree for the AtzD/barbiturase family. Clade 1 (red), containing predicted cyanuric acid hydrolases, is divided into three groups: group 1, Firmicutes enzymes; group 2, enzymes found in phylogenetically diverse bacteria with s-triazine degradation abilities (proteins sharing >99% identity to either AtzD [Pseudomonas sp. ADP, Pseudomonas nitroreducens, Comamonas sp. strain A2, Arthrobacter sp. strain MCMB-436, and Arthrobacter AD25] or TrzD [Acidovorax citrulli strain 12227, Enterobacter cloacae strain 99, and Aminobacter aminovorans] have condensed subtrees); and group 3, alphaproteobacterial enzymes. Clade 2 enzymes (blue) are found in actinobacteria and are divided into two groups, based on gene context data (see Fig. S1 in the supplemental material): group 4, containing an adjacent amino acid kinase superfamily protein, and group 5, containing the experimentally determined barbiturase from Rhodococcus erythropolis JCM 3132 and with an amidase signature superfamily member adjacent downstream. The outgroup from Frankia sp. Eul1c is in purple.
Table 1.
Cyanuric acid hydrolase/barbiturase family proteins identified in this study and identities to enzymes of known function
| Clade and organisma | GI no. | Locus name | % Identity to: |
|||
|---|---|---|---|---|---|---|
| AtzD | TrzD | Moorella cyanuric acid hydrolase | Barbiturase | |||
| 1 | ||||||
| Clostridium asparagiforme DSM 15981 | 225388419 | CLOSTASPAR_02155 | 51 | 57 | 65 | 45 |
| Moorella thermoacetica ATCC 39073 | 83590946 | Moth_2120 | 58 | 63 | 100 | 46 |
| Paenibacillus sp. JDR-2 | 251798893 | Pjdr2_4927 | 50 | 55 | 58 | 43 |
| Bacillus cellulosilyticus DSM 2522 | 317127947 | Bcell_1233 | 37 | 42 | 41 | 33 |
| Acidovorax citrulli strain 12227 | 3659839 | 58 | 100 | 63 | 45 | |
| Enterobacter cloacae strain 99 | 13022197 | 58 | 100 | 63 | 45 | |
| Aminobacter aminovorans (formerly Comamonas sp. A2) | 14029763 | Partial sequence | 58 | 100 | 63 | 45 |
| Bradyrhizobium japonicum USDA 110 | 27382392 | blr7281 | 59 | 55 | 59 | 42 |
| Pseudomonas sp. ADP | 32455884 | 100 | 58 | 58 | 43 | |
| Pseudomonas nitroreducens | 285310188 | 100 | 58 | 58 | 43 | |
| Comamonas sp. A2 | 254972448 | 100 | 58 | 58 | 43 | |
| Arthrobacter sp. MCMB-436 | 47059636 | 100 | 58 | 58 | 43 | |
| Arthrobacter AD25 | 117583156 | 99.7 | 58 | 58 | 43 | |
| Micromonas sp. strain RCC299 | 255079800 | MICPUN_59838 | 45 | 41 | 42 | 32 |
| Acidithiobacillus ferrooxidans ATCC 53993 and ATCC 23270 | 198283056 218519570 | Lferr_0922 AFE_0779 | 42 | 46 | 46 | 34 |
| Methylobacterium radiotolerans JCM 2831 | 170751160 | Mrad2831_4776 | 51 | 55 | 51 | 42 |
| Oceanicola granulosus HTCC2516 | 89068460 | OG2516_12979 | 44 | 44 | 44 | 37 |
| Agrobacterium vitis S4 | 222106665 | Avi_5653 | 48 | 50 | 56 | 43 |
| Rhizobium leguminosarum bv. viciae 3841 | 116254793 | pRL100353 | 48 | 52 | 54 | 43 |
| Rhizobium leguminosarum bv. trifolii WSM1325 | 241554234 | Rleg_6452 | 48 | 52 | 53 | 44 |
| Methylobacterium sp. 4-46 | 170741972 | M446_3816 | 46 | 47 | 47 | 37 |
| Bradyrhizobium sp. strain BTAi1 | 148252873 | BBta_1315 | 47 | 47 | 52 | 38 |
| Bradyrhizobium sp. strain ORS278 | 146343122 | BRADO6319 | 47 | 48 | 54 | 40 |
| Azorhizobium caulinodans ORS 571 | 158425516 | AZC_3892 | 50 | 52 | 55 | 41 |
| 2 | ||||||
| Nocardioides JS614 | 119718489 | Noca_4270 | 43 | 44 | 45 | 72 |
| Thermobispora bispora DSM 43833 | 296270184 | Tbis_2213 | 43 | 49 | 50 | 77 |
| Streptosporangium roseum DSM 43021 | 271967801 | Sros_6541 | 42 | 46 | 48 | 74 |
| Geodermatophilus obscurus DSM 43160 | 284988804 | Gobs_0181 | 42 | 46 | 45 | 80 |
| Catenulispora acidiphila DSM 44928 | 256391076 | Caci_1879 | 45 | 47 | 47 | 81 |
| Nakamurella multipartita DSM 44233 | 258655427 | Namu_5328 | 43 | 45 | 46 | 84 |
| Nocardioides sp. strain JS614 | 119715741 | Noca_1505 | 43 | 46 | 46 | 86 |
| Intrasporangium calvum DSM 43043 | 315588294 | Intca_1068 | 43 | 47 | 48 | 80 |
| Janibacter sp. strain HTCC2649 | 84497968 | JNB_17813 | 42 | 45 | 45 | 85 |
| Rhodococcus erythropolis JCM 3132 | 46395595 | 43 | 45 | 46 | 100 | |
| Rhodococcus erythropolis SK121 | 229318388 | RHOER0001_3381 | 43 | 45 | 46 | 99.7 |
| Rhodococcus erythropolis PR4 | 226309369 | RER_58820 | 43 | 45 | 46 | 99.7 |
| Rhodococcus opacus B4 | 226362213 | ROP_27990 | 42 | 46 | 45 | 92 |
| Rhodococcus jostii RHA1 | 111022263 | RHA1_ro05296 | 45 | 46 | 45 | 93 |
| Saccharomonospora viridis DSM 43017 | 257056105 | Svir_20970 | 44 | 46 | 47 | 85 |
| Azorhizobium caulinodans ORS 571 | 158424827 | AZC_3203 | 49 | 48 | 52 | 43 |
| Frankia sp. EuI1c | 280963003 | FraEuI1c_3137 | 36 | 36 | 40 | 38 |
Organisms containing homologs are in the same order as in the phylogenetic tree in Fig. 2. Organisms in which proteins were experimentally assayed are highlighted in bold.
The placement of proteins in four of the five groups correlated with the phylogeny of the source organisms. Groups 1 and 3 were composed exclusively of proteins from the Firmicutes and Alphaproteobacteria, respectively. Groups 4 and 5 were composed exclusively of actinobacterial proteins. These observations suggested that members of the cyanuric acid hydrolase/barbiturase family may be ancient, divergent, and chromosomally encoded in many organisms. The last point is supported by the gene placement within sequenced genomes (see Fig. S1 in the supplemental material).
Group 2 sequences were from a diverse set of organisms, representing Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Actinobacteria. All but one of the group 2 source organisms were isolated for their s-triazine degradation abilities, and the sequences from those organisms are >99% identical to either AtzD or TrzD (Fig. 2). Many of these genes have been associated with plasmids and transposons and could have spread to a variety of organisms via horizontal gene transfer (5, 7, 12, 26, 32). It is possible that some of these genes have spread more rapidly in recent times with the inadvertent selection pressure provided by the use of billions of pounds of anthropogenic s-triazine compounds.
Sequence alignments.
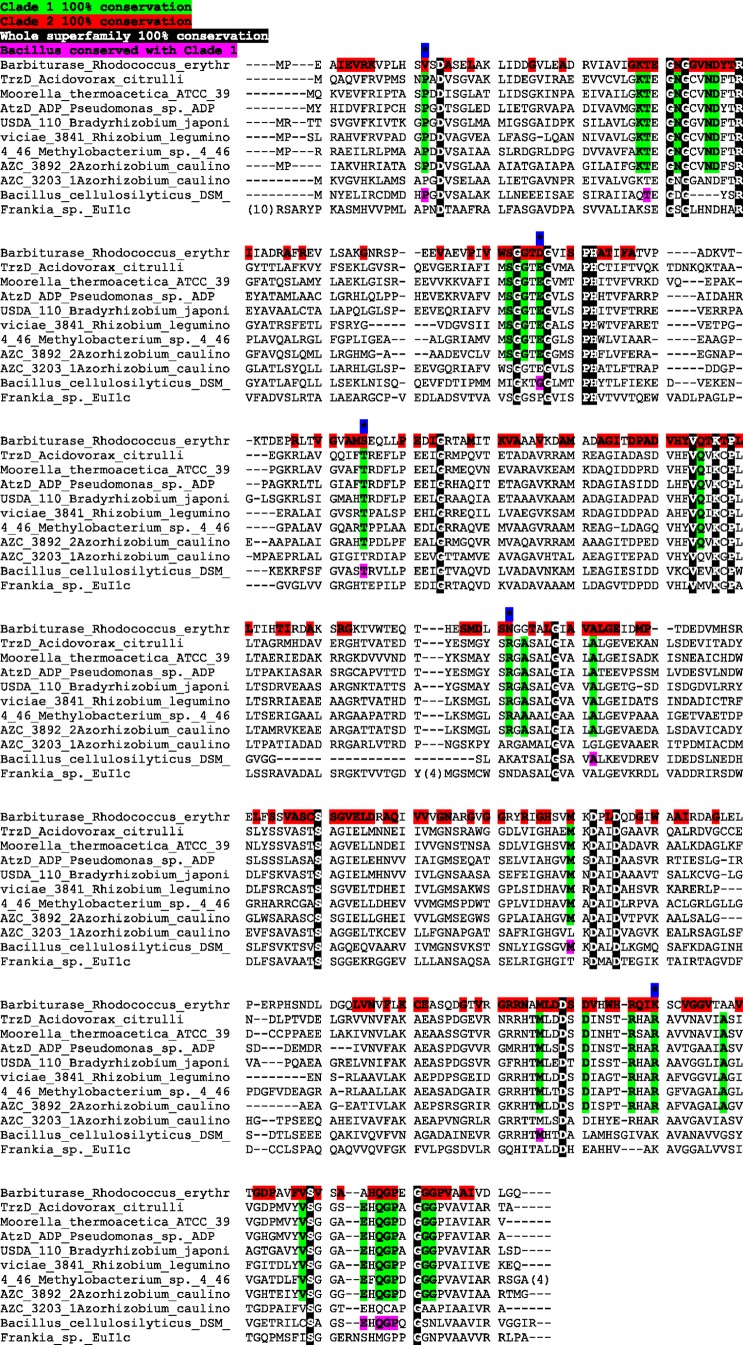
The degree of similarity across clade 2 is much greater than that of clade 1. The sequence conservation across the clades is represented in Fig. 3, which shows an alignment for all family members that have been experimentally characterized, either in this study or in studies previously reported in the literature. Those residues with 100% conservation across the entire family have a black background, while those conserved in clade 1 have a green background and those in clade 2 have a red background. Note that some residues were conserved in all but one sequence of the clade. These do not appear highlighted in the alignment. There are also positions that are conserved in all of clade 1 and have a different conserved residue in clade 2. These residues, indicated by an asterisk with a blue background, may be residues significant in substrate specificity between clades 1 and 2. A comprehensive family alignment can be found in Fig. S2 in the supplemental material. Further defining the catalytic and substrate specificity roles of residues conserved between and within each of the clades will require additional structural information (for example, that obtained from an X-ray structure determination).
Fig 3.
Sequence alignment of experimentally characterized and highly divergent cyanuric acid hydrolase/barbiturase family members. A black background indicates residues with 100% conservation over the entire family, a green background indicates 100% conservation across clade 1 (including sequences not in the current alignment), and a red background indicates 100% conservation across clade 2 (Note that only one sequence from clade 2 is shown in this alignment.) The blue background with an asterisk indicates different residues in clade 1 and clade 2 but 100% conservation within the respective clades.
Selection of enzymes for experimental investigation.
The three cyanuric acid hydrolases with an experimentally determined function are located in group 1 (from Moorella thermoacetica ATCC 39073) and group 2 (AtzD and TrzD). None of the group 3 proteins had been functionally characterized until this study. The only known functional protein in clade 2 was barbiturase from Rhodococcus erythropolis JCM 3132, which was placed into group 5. To expand the knowledge of the functional sequence space in the family, five of the newly identified proteins were selected for further study, as described in the next section. The proteins were chosen either to represent a group with no functionally characterized members or as atypical proteins from groups containing characterized proteins.
The Bradyrhizobium japonicum USDA 110 protein was chosen due to its unusual placement in the tree compared to other Bradyrhizobium and Rhizobium strains. The Methylobacterium sp. 4-46 protein was one of 3 proteins chosen to represent group 3. The Rhizobium leguminosarum bv. viciae 3841 protein was chosen as another group 3 member because it was plasmid encoded. Azorhizobium caulinodans ORS 571 was found to have two family member proteins. The protein encoded by locus AZC_3892 was placed into group 3, while the protein encoded by locus AZC_3203 was the only potential family member, apart from the outgroup (the sequence from Frankia sp. Eul1c), that was not placed into clade 1 or 2 (Fig. 2). Both Azorhizobium proteins were selected for further characterization to determine if they had the same or different functionalities. No clade 2 proteins were chosen for further characterization due to the limited diversity among the sequences. The genes for each of these AtzD/barbiturase family homologs were PCR amplified, cloned, and expressed in E. coli, and the resulting proteins were purified and assayed.
Enzymatic activity and kinetic determinations.
The clustering of the Bradyrhizobium japonicum USDA 110 protein with the AtzD protein from Pseudomonas sp. ADP (12) suggested that this enzyme was a cyanuric acid hydrolase. However, with less than 59% identity to any of the experimentally confirmed cyanuric acid hydrolases, this functionality was ambiguous. Experimental studies of the recombinant protein confirmed it to be a cyanuric acid hydrolase. Furthermore, the proteins from group 3 of the phylogenetic tree, Methylobacterium sp. 4-46, Rhizobium leguminosarum bv. viciae 3841, and Azorhizobium caulinodans ORS 571 locus AZC_3892, were also confirmed to have cyanuric acid hydrolase activity. These are the first functional assignments to this section of the tree, and they show that the cyanuric acid hydrolase enzymes are distributed throughout clade 1. In addition, the Bradyrhizobium japonicum USDA 110 and Azorhizobium caulinodans ORS 571 locus AZC_3892 proteins were shown not to be reactive with barbituric acid. The second homolog from Azorhizobium caulinodans ORS 571, locus AZC_3203, failed to display either cyanuric acid hydrolase or barbiturase activity, consistent with its position outside both clades 1 and 2 (Fig. 2). The AZC_3203 protein was further tested for activity with other potential substrates, including uracil, thymine, 6-azauracil, 5-azacytosine, cytosine, 2,4,5-trihydroxypyrimidine, 5-nitrobarbaturic acid, creatinine, 5,6-dihydroxyuracil, hydantoin, ammelide, ammeline, allantoin, alloxan, urea, l-asparagine, and biuret. No hydrolysis of any of these compounds could be detected by spectroscopy or HPLC.
Steady-state kinetic constants were determined to investigate whether those parameters would indicate that cyanuric acid hydrolysis was a major physiological function for these proteins. In general, physiological enzymes show kcat/Km ratios of at least 104 M−1 s−1 for their preferred substrates. For comparative purposes, AtzD from Pseudomonas sp. ADP and TrzD from Acidovorax citrulli strain 12227 were also purified and kinetic values determined. Note that the AtzD protein has been demonstrated to be part of an operon that is regulated partly by cyanuric acid (13) and that TrzD was isolated from a bacterium that grows on cyanuric acid as a sole nitrogen source.
Stability studies with the Pseudomonas sp. ADP enzyme found that when the enzyme was frozen, a substantial loss in activity occurred. However, storage of the enzyme at 4°C resulted in stable activity for more than 1 month. For this reason, all enzymes were stored under these new conditions and activity monitored throughout the study to ensure no detectable loss of activity. The kcat for the Pseudomonas sp. ADP enzyme, stored in this way, was 10-fold higher than that previously published (11). However, purified TrzD had a kcat that was 10-fold lower than that previously published (20). The kcat and Km values for the newly cloned cyanuric acid hydrolases were each within an order of magnitude of each other (Table 2). The kcat values ranged from 5 to 73 s−1 and the Km values from 23 to 370 μM. These give kcat/Km values that ranged from 4 × 104 to 5 × 106. These values suggested that cyanuric acid hydrolysis is the physiological function of these enzymes. In contrast, the published values for the barbiturase are lower for kcat and higher for Km. Thus, the kcat/Km is lower (Km = 1 mM, kcat = 1.6 s−1, and kcat/Km = 1.6 × 103) (34). However, the genome and physiological context indicate that barbituric acid hydrolysis is the physiological function for this enzyme.
Table 2.
Kinetic constants of various cyanuric acid hydrolase/barbiturase family enzymes, using cyanuric acid as a substrate
| Enzyme name and /or organism (reference) | kcat (s−1) | Km (μM) | kcat/Km (s−1 M−1) |
|---|---|---|---|
| TrzD (20) | 250 | 50 | 5 × 106 |
| TrzD (this study) | 14.2 ± 0.4 | 58 ± 7 | (2.5 ± 0.4) × 105 |
| Pseudomonas sp. ADP AtzD (12) | 6.8 ± 0.7 | 57 ± 10 | (1.2 ± 0.3) × 105 |
| Pseudomonas sp. ADP AtzD (this study) | 73 ± 6 | 23 ± 7 | (3.2 ± 1.2) × 106 |
| Moorella thermoacetica ATCC 39073 (23) | 10.6 | 110 | 1.0 × 105 |
| Bradyrhizobium japonicum USDA 110 | 9.3 ± 0.7 | 50 ± 10 | (1.9 ± 0.5) × 105 |
| Rhizobium leguminosarum bv. viciae 3841 | 5 ± 1 | 130 ± 60 | (3.8 ± 2.5) × 104 |
| Methylobacterium sp. 4-46 | 17 ± 2 | 69 ± 16 | (2.5 ± 0.9) × 105 |
| Azorhizobium caulinodans ORS 571 locus AZC_3892 | 50 ± 9 | 370 ± 90 | (1.3 ± 0.6) × 105 |
| Azorhizobium caulinodans ORS 571 locus AZC_3203 | —a | — | — |
—, no activity with any substrate tested.
Demonstration that cyanuric acid hydrolases and barbiturases are isofunctional with their respective substrates.
Based on overall sequence relatedness and multiple-sequence alignments, it seemed odd that barbiturases and cyanuric acid hydrolases would catalyze different reactions. Biuret had been reported as the product of the cyanuric acid hydrolase reaction (8, 12, 20, 23), including in one previous study using 13C NMR (36). However, in that previous 13C NMR experiment, the reaction mixture was analyzed at a pH of 4.0, a pH at which carboxybiuret, if formed, would be highly unstable. In that context, this experiment was reexamined here.
In the present study, 13C NMR was used to analyze reaction mixtures poised at pH 7.7. Cyanuric acid is a symmetrical molecule (Fig. 1A), and the three carbon atoms appeared as a single resonance (157.7 ppm) in the 13C NMR spectrum, as expected (Fig. 1A and 4A). Biuret is also symmetrical (Fig. 1A), and carbonate/bicarbonate contains a single carbon atom; therefore, single resonances were expected. Experiments confirmed this, with chemical shifts of 157.5 and 160.4 ppm, respectively. The three carbon atoms in the proposed carboxybiuret intermediate would be bonded to different substituents (Fig. 1A) and would be expected to appear as three separate resonances. Prior to enzyme addition, a stable signal for [U-13C]cyanuric acid was observed at 157.7 ppm (Fig. 4A). The addition of cyanuric acid hydrolase from Moorella thermoacetica ATCC 39073 led to a decrease in the cyanuric acid resonance of approximately 40% and, concomitantly, showed three new signals (157.1 ppm, 156.8 ppm, and 155.0 ppm), which we attributed to the presence of carboxybiuret (Fig. 4B). No bicarbonate/carbonate signal was observed at this time. As the reaction proceeded (Fig. 4C), a peak attributed to bicarbonate/carbonate appeared at 160.4 ppm. The resonance(s) at 157.5 to 155.7 ppm became broad and misshapen due to the appearance of biuret carbon atoms resonating at 157.5 ppm and overlapping with the residual cyanuric acid peak at 157.7 ppm. As the reaction continued, the cyanuric acid peak shifted slightly upfield, likely due to increased acidity of the reaction mixture. The biuret peak increased and sharpened (157.5 ppm) (Fig. 4D). Concurrently, the bicarbonate/carbonate peak also increased. At this stage of the reaction, the integration of the bicarbonate/carbonate-biuret signals yielded a molar stoichiometry of 1:1, as expected. Also, the ratio of the proposed carboxybiuret carbon resonances was 1:1:1, as expected. (Fig. 4D). Within 2 h, only bicarbonate/carbonate and biuret were detected (Fig. 4E). Formation of carboxybiuret via 13C NMR was observed with the cyanuric acid hydrolases from Pseudomonas sp. ADP, Moorella thermoacetica ATCC 39073, and Azorhizobium caulinodans ORS 571 (locus AZC_3892). These results suggested that cyanuric acid hydrolases uniformly catalyze a hydrolytic amide bond cleavage that is analogous to the barbiturase reaction (Fig. 1).
Fig 4.
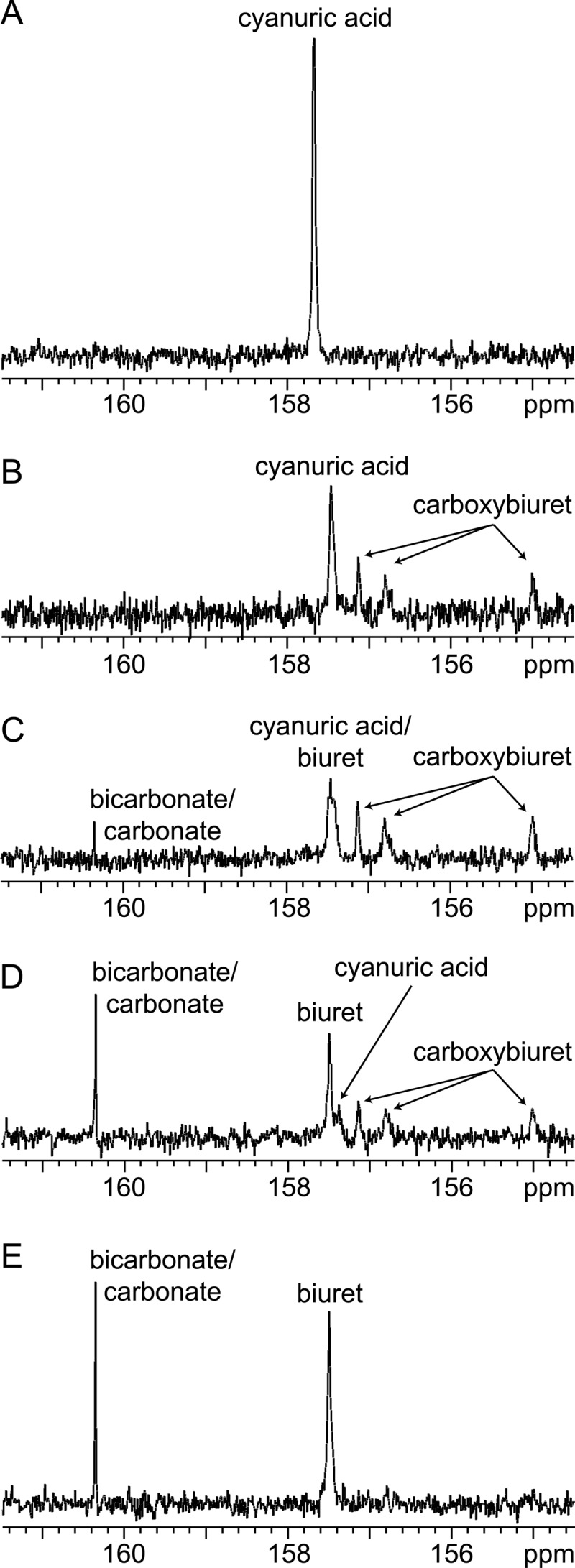
13C NMR of cyanuric acid hydrolase biotransforming [U-13C]cyanuric acid to [U-13C]carboxybiuret, followed by nonenzymatic decarboxylation. (A) [U-13C]cyanuric acid without enzyme; (B to E) [U-13C]cyanuric acid with enzyme at 9 min (B), 14 min (C), 41 min (D), and 112 min (E).
Attempts to synthesize carboxybiuret were unsuccessful, and, to our knowledge, its synthesis has not been described in the literature. In light of this, we also employed mass spectrometry with electrospray ionization to support the identification of carboxybiuret. Cyanuric acid with natural isotopic abundance was incubated with cyanuric acid hydrolase and observed over time. A parallel experiment was performed using [U-13C]cyanuric acid. In negative-ion mode, we observed mass ions consistent with the formation, and subsequent decomposition, of carboxybiuret adducts over the time course. Two pairs of ions were examined in detail. Each pair contained one ion from the experiment with unlabeled cyanuric acid and a corresponding heavier ion from the experiment with [U-13C]cyanuric acid. In the case of the ion pair with the nominal masses of 244 and 247 Da, these were attributed to carboxybiuret combining with potassium acetate (see Fig. S3 in the supplemental material). In the case of the ion pair with masses of 347 and 352 Da, these could be generated from carboxybiuret in complexation with biuret and potassium acetate or phosphoric acid. We did not observe the parent ion of carboxybiuret. This is likely because it was too unstable to survive the electrospray or the mass spectrometer analyzer and thus was seen only when complexed with another ion that provided some protection. In total, the mass spectrometry data supported the conclusion that carboxybiuret is the intermediate generated by cyanuric acid hydrolase.
Evidence against the AtzD/barbiturase family linkage to the amidohydrolase superfamily.
Initially, the barbiturase from Rhodococcus erythropolis JCM 3132 was identified as a member of the amidohydrolase superfamily due to the presence of a C-terminal HxH site in the protein sequence and a 1:1 zinc-to-subunit ratio (34). The homologous AtzD protein, however, lacked any observable bound metal (12). The current study further establishes that, beyond these two proteins being homologous, the reactions that they catalyze are completely analogous. Having homologous proteins catalyzing analogous reactions with two different mechanisms, one involving a metal and one without, is highly unusual, prompting additional investigation into this matter.
Extensive sequence comparisons were conducted here to assess AtzD/barbiturase family linkages or their absence. Results from pblast, PSI-BLAST, Shotgun, and HMMER were unable to link any AtzD/barbiturase family member to an outside family or superfamily, including the amidohydrolase superfamily. Likewise, analysis of over 33,000 amidohydrolase sequences found in the Structure Function Linkage Database (29) failed to connect to any member of the AtzD/barbiturase family.
The amidohydrolase proteins comprise an extensive superfamily, usually containing one or two metals that are liganded by a set of conserved histidines and, in many cases, carboxylic acid residues. The amino acid ligands span the length of the linear protein sequence, with the HxH site being the first, located in the N-terminal part of the protein. The protein folds into a (β/α)8 barrel in which the metal ligands, located in conserved positions within the secondary structure, converge into the center of the barrel to coordinate the metal (33). Secondary-structure predictions for the AtzD/barbiturase family were not able to establish similarity in structure or to find metal ligands comparable to those in the amidohydrolase superfamily. The only commonality between this superfamily and the barbiturase is the HxH site. The presence of an HxH site alone is not enough to link proteins to the amidohydrolase superfamily, especially since the amidohydrolase site is in the N terminus and the barbiturase site is in the C terminus of the protein. Countless other proteins also have an HxH site without being related to the amidohydrolase superfamily (6, 17, 19, 27, 38). Furthermore, analysis of the AtzD/barbiturase family sequences reveals that this HxH site, initially identified in a single protein from Rhodococcus erythropolis JCM 3132, is not completely conserved (Fig. 3). If this motif were required for metal binding and catalysis, conservation across the entire protein family would be expected.
Previous studies on enzymes from Acidovorax citrulli strain NRRLB-12227, Pseudomonas sp. ADP, and Moorella thermoaceticaATCC 39073 found no stimulation from adding divalent metal ions (12, 20, 23). However, in the present study, we did analyze for the presence or absence of a stoichiometrically bound metal. The cyanuric acid hydrolases from Pseudomonas sp. ADP, Moorella thermoacetica ATCC 39073, and Bradyrhizobium japonicum USDA 110 and the barbiturase from Rhodococcus erythropolis JCM 3132 were subjected to metal analysis under the same conditions. Zinc, iron, cobalt, nickel, copper, manganese, magnesium, lead, cadmium, and chromium were looked for. Only zinc and nickel were above background levels, and the amounts of those metals combined accounted for less than 0.1 metal per subunit. This differs greatly from the case for amidohydrolase superfamily members, which usually have 1 to 2 metals per subunit, as determined in our laboratory by this same method. These data suggested that both the cyanuric acid hydrolases and barbiturase are not metalloproteins.
DISCUSSION
This study is the first to describe the cyanuric acid hydrolases and barbiturases as a protein family. Previously, the barbiturases were suggested to be zinc enzymes in the amidohydrolase superfamily, but sequence analysis and metal determination conducted here argue against that family assignment and place them, along with the cyanuric acid hydrolases, in a unique protein family. This study further established that both enzymes catalyze common chemistry: a hydrolytic ring amide bond cleavage reaction, resulting in the opening of a heterocyclic 6-membered ring. The NMR and mass spectrometry experiments conducted here show for the first time that cyanuric acid hydrolases release carboxybiuret and not biuret.
The known set of proteins in this protein family was expanded from 4 to 41. Still, this is a rare family, appearing on an average of once in every 200 sequenced genomes (from a set of 6,423 genomes). The lack of a crystal structure within this set prevents the identification of the fold. Fold recognition/homology modeling programs such as SUPERFAMILY (15), pGenTHREADER (25), pDomTHREADER (25), Phyre2 (21), and SWISS-MODEL (1) were unable to provide any insights into structural classification. However, sequence comparisons, followed up by purification and activity measurements, allowed us to differentiate barbiturases from cyanuric acid hydrolases. The latter are the larger set of proteins known at this time, mainly due to their prevalence in s-triazine-degrading bacteria. The mobility of s-triazine degradation genes and high sequence identity of this group (group 2 in Fig. 2) suggest that these proteins may be part of a resurgence of cyanuric acid hydrolases due to the large amount of anthropomorphic s-triazine chemicals being produced currently. s-Triazine herbicides and melamine alone are produced in quantities greater than one hundred million pounds and one billion pounds, respectively (22).
Cyanuric acid is found naturally, but its occurrence derives from spontaneous rather than metabolic chemistry. Cyanuric acid has been recovered from meteorites (16), and s-triazines have been identified in “primordial soup” experiments (18). Cyanuric acid also is formed intracellularly during the radiative decay of purine nucleotides. These observations are consistent with the data in this paper that suggest an ancient set of genes without a strong selection pressure for their prevalence.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Biocatalysis and Synthetic Ecology Initiatives of the BioTechnology Institute at the University of Minnesota (to L.P.W. and M.J.S.), a grant from the National Center for Food Protection and Defense, and a fellowship from the NIH (Biotechnology Training Grant GM08347) (to S.C.).
We thank Steve Harvey at The Center for Mass Spectrometry and Proteomics facility, University of Minnesota, St. Paul, MN, for assistance with mass spectrometry.
Footnotes
Published ahead of print 22 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 2. Brown SD, Gerlt JA, Seffernick JL, Babbitt PC. 2006. A gold standard set of mechanistically diverse enzyme superfamilies. Genome Biol. 7:R8 doi:10.1186/gb-2006-7-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chubb D, Jefferys BR, Sternberg MJ, Kelley L. 2010. Sequencing delivers diminishing returns for homology detection: implications for mapping the protein universe. Bioinformatics 26:2664–2671 [DOI] [PubMed] [Google Scholar]
- 4. Copley S. 2009. Prediction of function in protein superfamilies. F1000 Biol. Rep. 1:91 doi:10.3410/B1-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Souza ML, Wackett LP, Sadowsky MJ. 1998. The atzABC genes encoding atrazine catabolism are located on a self-transmissible plasmid in Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 64:2323–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26:740–746 [DOI] [PubMed] [Google Scholar]
- 7. Eaton RW, Karns JS. 1991. Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J. Bacteriol. 173:1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eaton RW, Karns JS. 1991. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J. Bacteriol. 173:1363–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
- 10. Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 11. Fruchey I. 2001. Purification and characterization of AtzD: a novel cyanuric acid amidohydrolase from Pseudomonas sp. strain ADP. Master's thesis University of Minnesota, St. Paul, MN [Google Scholar]
- 12. Fruchey I, Shapir N, Sadowsky MJ, Wackett LP. 2003. On the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 69:3653–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. García-González V, Govantes F, Porrúa O, Santero E. 2005. Regulation of the Pseudomonas sp. strain ADP cyanuric acid degradation operon. J. Bacteriol. 187:155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glasner M, Gerlt J, Babbitt P. 2006. Evolution of enzyme superfamilies. Curr. Opin. Chem. Biol. 10:492–497 [DOI] [PubMed] [Google Scholar]
- 15. Gough J, Chothia C. 2002. Superfamily: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 30:268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayatsu R, Studier MH, Oda A, Fuse K, Anders E. 1968. Origin of organic matter in early solar system. II. Nitrogen compounds. Geochim. Cosmochim. Acta 32:175–190 [Google Scholar]
- 17. Hooper NM. 1994. Families of zinc metalloproteases. FEBS Lett. 354:1–6 [DOI] [PubMed] [Google Scholar]
- 18. Hysell M, Siegel JS, Tor Y. 2005. Synthesis and stability of exocyclic triazine nucleosides. Org. Biomol. Chem. 3:2946–2952 [DOI] [PubMed] [Google Scholar]
- 19. Karlin S, Zhu Z. 1997. Classification of mononuclear zinc metal sites in protein structures. Proc. Natl. Acad. Sci. U. S. A. 94:14231–14236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karns JS. 1999. Gene sequence and properties of an s-triazine ring-cleavage enzyme from Pseudomonas sp. strain NRRLB-12227. Appl. Environ. Microbiol. 65:3512–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kelley L, Sternberg M. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 22. LeBaron H, McFarland J, Burnside O. 2008. The triazine herbicides. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 23. Li Q, Seffernick JL, Sadowsky MJ, Wackett LP. 2009. Thermostable cyanuric acid hydrolase from Moorella thermoacetica ATCC 39073. Appl. Environ. Microbiol. 75:6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides N. 2006. The genomes on line database (GOLD) v. 2: a monitor of genome projects worldwide. Nucleic Acids Res. 34:D332–D334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lobley A, Sadowski M, Jones D. 2009. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 25:1761–1767 [DOI] [PubMed] [Google Scholar]
- 26. Martinez B, Tomkins J, Wackett LP, Wing R, Sadowsky MJ. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Odintsov SG, Sabala I, Marcyjaniak M, Bochtler M. 2004. Latent LytM at 1.3 Å resolution. J. Mol. Biol. 335:775–785 [DOI] [PubMed] [Google Scholar]
- 28. Pegg SC, et al. 2005. Representing structure-function relationships in mechanistically diverse enzyme superfamilies. Pac. Symp. Biocomput. 2005:358–369 [PubMed] [Google Scholar]
- 29. Pegg SC, et al. 2006. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry 45:2545–2555 [DOI] [PubMed] [Google Scholar]
- 30. Pegg SC, Babbitt PC. 1999. Shotgun: getting more from sequence similarity searches. Bioinformatics 15:729–740 [DOI] [PubMed] [Google Scholar]
- 31. Seffernick JL, Johnson G, Sadowsky MJ, Wackett LP. 2000. Substrate specificity of atrazine chlorohydrolase and atrazine-catabolizing bacteria. Appl. Environ. Microbiol. 66:4247–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seffernick JL, Wackett LP. 2001. Rapid evolution of bacterial catabolic enzymes: a case study with atrazine chlorohydrolase. Biochemistry 40:12747–12753 [DOI] [PubMed] [Google Scholar]
- 33. Seibert CM, Raushel FM. 2005. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44:6383–6391 [DOI] [PubMed] [Google Scholar]
- 34. Soong CL, Ogawa J, Sakuradani E, Shimizu S. 2002. Barbiturase, a novel zinc-containing amidohydrolase involved in oxidative pyrimidine metabolism. J. Biol. Chem. 277:7051–7058 [DOI] [PubMed] [Google Scholar]
- 35. Soong CL, Ogawa J, Shimizu S. 2001. Novel amidohydrolytic reactions in oxidative pyrimidine metabolism: analysis of the barbiturase reaction and discovery of a novel enzyme, ureidomalonase. Biochem. Biophys. Res. Commun. 286:222–226 [DOI] [PubMed] [Google Scholar]
- 36. Stamper D, Krzycki J, Nicomrat D, Traina S, Tuovinen O. 2005. Ring-cleaving cyanuric acid amidohydrolase activity in the atrazine-mineralizing Ralstonia basilensis M91-3. Biocatal. Biotransform. 23:387–396 [Google Scholar]
- 37. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woo E, et al. 2002. Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 21:2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.