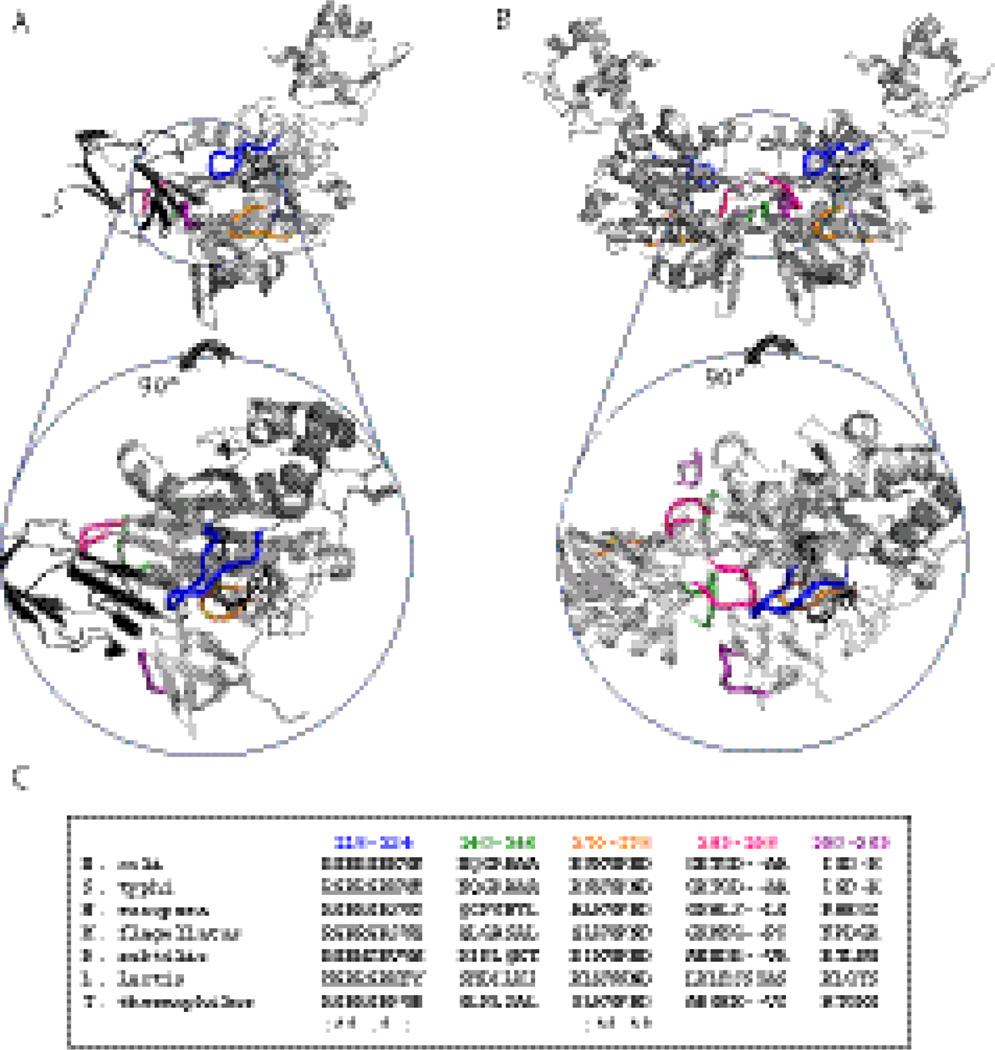
Fig. 2.
Models of holoBirA-BCCP87 heterodimer (A) and holoBirA homodimer (B). Rotation of the molecules by 90° displays cross-sections of the heterodimer interface (bottom left) and homodimer interface (bottom right), highlighting the surface loops involved in the two interactions. All models were created using Pymol39 with input file 2EWN for the homodimer and a file constructed by Zachary Wood for the heterodimer. This model, which uses the experimentally determined structure of the Pyroccocushorikoshi biotin protein ligase:BCCP complex as a template (2EJG)19, was constructed using the holoBirA monomer coordinates from 2EWN12 and the coordinates for apoBCCP87 (1BDO)40 C. Multiple sequence alignment of several bifunctional microbial biotin protein ligases performed using Clustal W.16 The color codes for loop sequences correspond to the coloring of loops in the models in A and B.