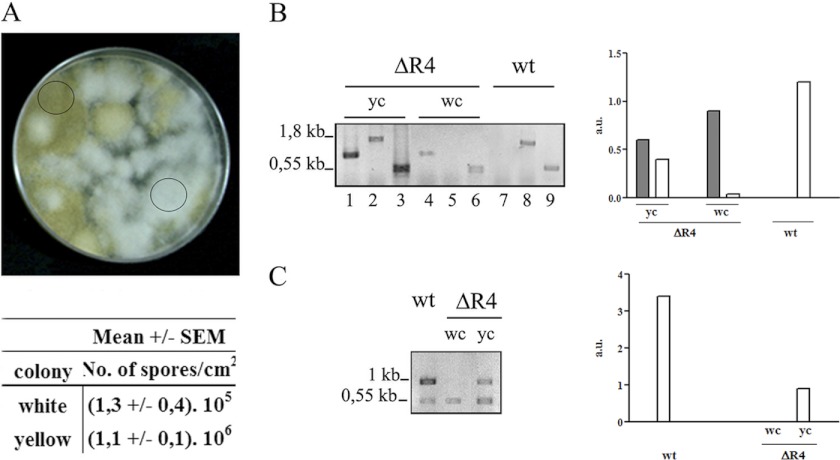
Fig 2.
Heterokaryotic ΔR4 strain. (A) ΔR4 strain grown in the selective medium MMC (pH 4.5) in triplicate and incubated at 30°C for 5 days. The circles indicate a white colony and a yellow colony. The numbers of spores per cm2 from white and yellow colonies were determined. Plugs of agar (1 cm2) were removed in triplicate for spore calculation. (B) PCR performed with genomic DNA from the wild-type strain (wt), a yellow colony (yc), and a white colony (wc) with the primers pkaR4F-Pr (lanes 1, 4, and 7) or pkaR4F-pkaR4R (lanes 2, 5, and 8). Tef-1f-Tef-1r was used as an internal control (lanes 3, 6 and 9). The primers are shown in Fig. 1E. (C) RT-PCR performed with RNA from wt, dc and wc with primers pkaR4F-pkaR4R, with Tef-1f/Tef-1r as internal control. The positions and sizes of the DNA fragments are indicated at the left of the panels. The quantification of the PCR products, after normalization with the tef-1 signal, is shown in the right panel. The dark bars represent the mutant allele, and the white bars represent the wt allele. The primers used are shown in Fig. 1E.