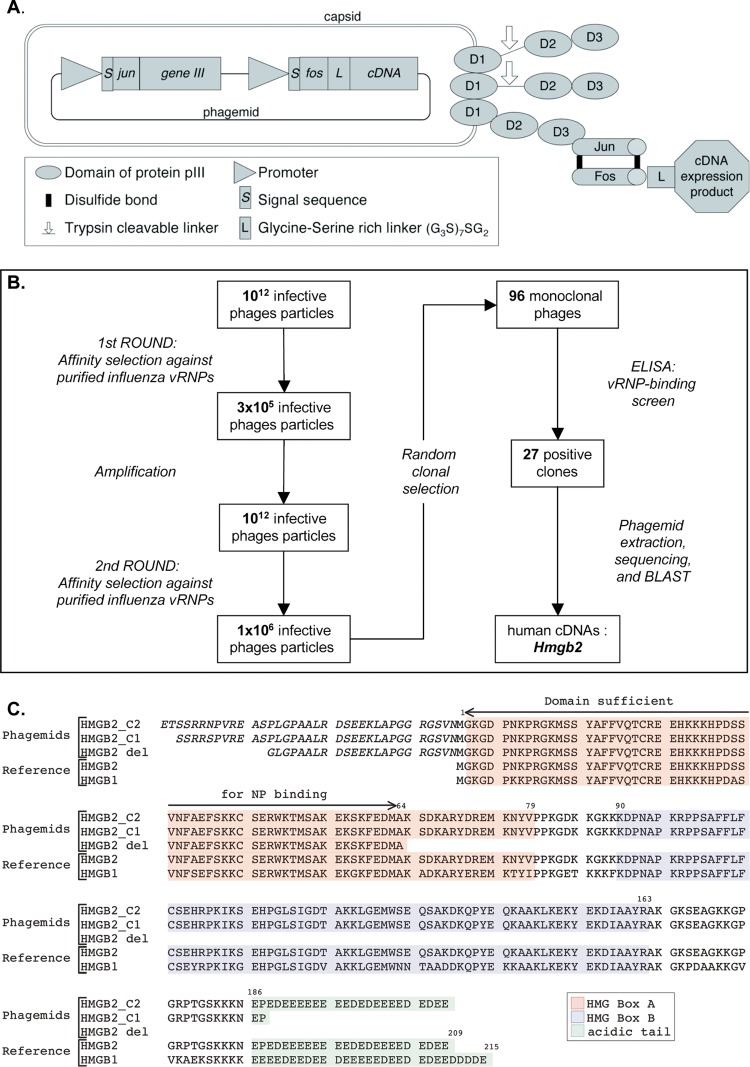
Fig 1.
Selection of influenza virus RNP-binding proteins by phage display. (A) Phage display system. The phages contain a single-stranded phagemid DNA packaged into a filamentous particle that contains three to five copies of the minor coat protein pIII at one of its tips. The three domains of pIII (D1, D2, and D3) are represented schematically. The pIII proteins derived from the KM13 helper phage contain a protease-cleavable linker between domains D2 and D3 (indicated by an arrow). The pIII proteins encoded by the phagemid are fused at their N-terminal ends to the Jun peptide. The Fos peptide is at the N terminus of a glycine/serine-rich linker (G3S)7SG2 which is fused to the human cDNA expression product encoded by the phagemid. The Fos and Jun peptides are bound by two disulfide bonds (black lines). (B) Schematic of the serial affinity selection cycles and monoclonal phage-protein screen. (C) Human cDNA sequences identified upon phage display. Two human HMGB2-derived sequences selected by affinity for viral RNPs (HMGB2 C1 and C2) and the truncated human HMGB2-del sequence are shown, together with the reference amino acid sequences of the human HMGB2 (gene ID 3148) and HMGB1 (gene ID 3146) proteins. HMGB2 and HMGB1 share two DNA-binding domains named HMG box A (highlighted in red) and HMG box B (highlighted in blue), as well as an acidic tail (highlighted in green). The residues indicated in italic derive from the 5′ noncoding region of the cDNA inserts, in the context of the Fos peptide-linker-cDNA expression product fusion.