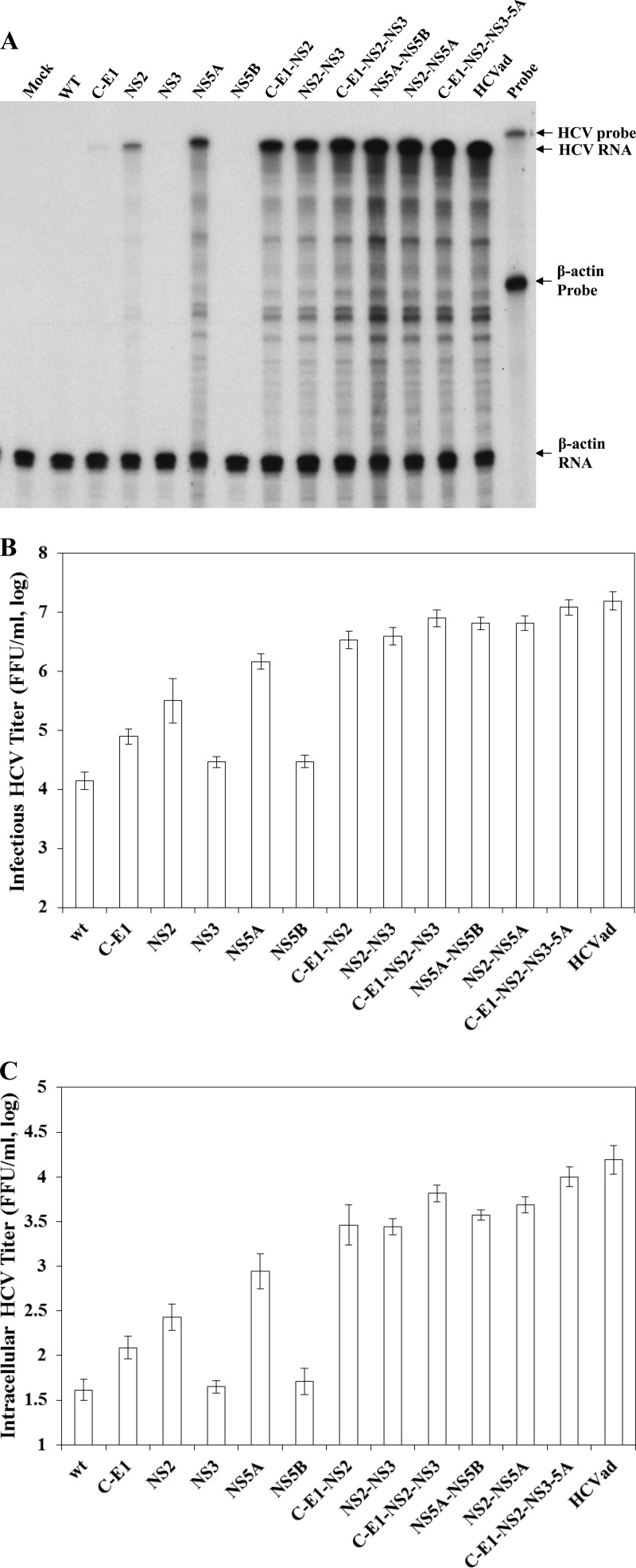
Fig 3.
Effects of adaptive mutations in different combinations on HCV production. Infectious HCV RNAs containing adaptive mutations in HCV proteins (indicated on the top) were transfected into Huh-7.5 cells using the DMRIE-C reagent following the manufacturer's instructions. At 72 h p.t., total cellular RNAs were extracted with TRIzol reagent, while the cell culture supernatants were collected and used for the determination of HCV infectivity in the supernatants. Upon infection with HCV in the supernatants, the total RNAs in the infected Huh-7.5 cells were extracted using TRIzol reagent. The levels of HCV positive-strand RNA in the HCV-infected naive Huh-7.5 cells were determined by RPA (A). Both infectious HCV titers in the medium (B) and in the cells (C) were determined by serial dilution and IFA, as described previously. Mean values and standard deviations shown in the graph are derived from three independent experiments. The enhancement of infectious HCV production by combination of adaptive mutations in C-E1, NS3, and NS5B with that in NS2 or those in NS5A was statistically significant (P < 0.05) compared with the NS2 or NS5A mutations per se or highly significant (P < 0.01) compared with wild-type virus or adaptive mutations in C-E1, NS3, and NS5B alone.