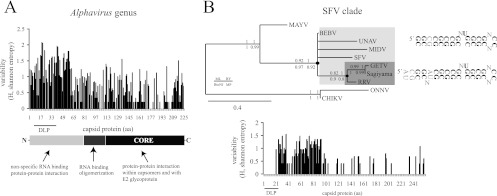
Fig 4.
Some evolutionary patterns of DLPs in Alphavirus. (A) Flexibility of the N-terminal region of the capsid protein to changes allowed the appearance of DLP structures in the alphavirus. The variability in capsid protein sequences among representative alphaviruses was calculated by the Shannon entropy method as described in Materials and Methods. The domain organization of the SINV prototype of the capsid protein is shown. DLP structures are localized in the coding regions of 26S mRNA corresponding to amino acids (aa) 7 to 40 of the capsid protein. (B) Conservation of DLP structure and the corresponding amino acid sequence among species of the SFV clade (SFV, BEBV, UNAV, MIDV, GETV, Sagiyama virus, and RRV). Shown is a phylogenetic tree of the SFV clade based on the first 150 nt of capsid coding sequences that include the DLP structures. Phylogenetic analysis was carried out by the maximum likelihood method using the GTR+I+Γ substitution model. Neighbor-joining (BioNJ), Bayesian (MrBayes), and maximum parsimony (MP) analysis resulted in an almost equivalent tree topology. The numbers shown are the branch support by 1,000 bootstrap resamplings for BioNJ, PhyML, and MP and the posterior probabilities for MrBayes, as indicated in the legend. Branches with <60% support value were collapsed. According to this model, a stable DLP may have emerged at the base of SFV/BEBV/UNAV/MIDV/GETV/Sagiyama virus/RRV clade that further evolved in the RRV/GETV/Sagiyama virus branch (denoted by black dots). The conserved nucleotide framework of DLPs is in bold, whereas the variable positions among viral species of the same clade are denoted by “N.” The variability in capsid protein sequences (Shannon entropy) among members of the SFV/BEBV/UNAV/MIDV/GETV/Sagiyama virus/RRV complex is shown in the lower panel.