Fig 3.
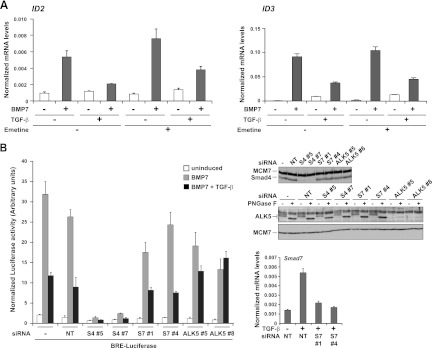
Inhibition of BMP7 responses by TGF-β does not require new protein synthesis and is independent of Smad7. (A) MDA-MB-231 cells were pretreated for 10 min with or without 400 μM emetine and then induced with the ligands indicated for 1 h. Total RNA was prepared, and levels of ID2 or ID3, relative to that of the gene for glyceraldehyde-3-phosphate dehydrogenase, were measured by qPCR. The qPCRs were performed in triplicate, and means and standard deviations are shown. (B) MDA-MB-231 BRE cells were transfected with the indicated siRNAs or a nontargeting (NT) control for 72 h before being treated with the ligands indicated for 8 h. Luciferase/Renilla assays were performed as described in the legend to Fig. 1. The knockdown efficiency of the Smad4 (S4) and ALK5 siRNAs is shown by Western blotting. For the ALK5 blot assay, extracts were treated with or without peptide N-glycosidase F (PNGase F) (12) to remove sugars from ALK5, enabling it to be more easily detected. The efficiency of the Smad7 (S7) knockdown was established by qPCR. Samples were treated for 1 h with TGF-β to induce levels of Smad7 in the control samples to be detectable by qPCR. Levels of Smad7, relative to that of the gene for glyceraldehyde-3-phosphate dehydrogenase, were calculated. Means and standard deviations of a representative experiment performed in triplicate are shown.
