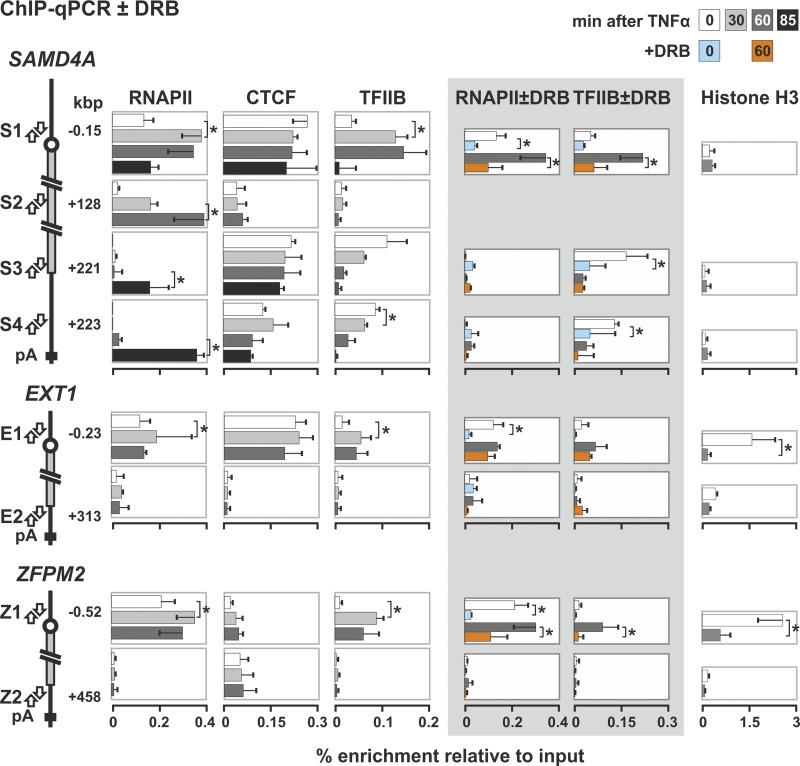
Fig 6.
Changes induced by TNF-α on the binding of RNA polymerase II, CTCF, TFIIB, and nucleosomes to each end of SAMD4A, EXT1, and ZFPM2. HUVECs were treated with TNF-α for the times indicated, and ChIP was performed with antibodies against the largest subunit of RNA polymerase II, CTCF, TFIIB, and histone H3 (percent enrichments ± the SD; n = 6). The diagrams on the left illustrate targets of the primer pairs in promoters (S1, E1, and Z1), the SAMD4A gene body (S2), and 3′ untranslated regions (UTRs; S3, S4, E2, and Z2). The polymerase is found at all promoters, both before and after stimulation. It is also seen at the 3′UTR of SAMD4A after 85 min (as expected), but not at the 3′UTR of EXT1 or ZFPM2 (since pioneering polymerases only reach the end of these two long genes after the end of the experiment). CTCF binds to promoters throughout the time course with comparable enrichments. It also binds to S3 and S4 but not to S2, E2 or Z2. TFIIB binding to all promoters peaks after 10 to 30 min. Some TFIIB binds to the 3′UTR of SAMD4A but not to the middle of SAMD4A (as expected) or the 3′UTRs of EXT1 or ZFPM2. When 50 μM DRB is added to cells 25 min before harvesting (gray panel), less TFIIB binds to the SAMD4A 3′UTR. H3 levels are generally low, except for E1 and Z1 at 0 min, which changes on stimulation (perhaps as the nucleosomes reposition). *, difference significant (P < 0.01, two-tailed unpaired Student t test).