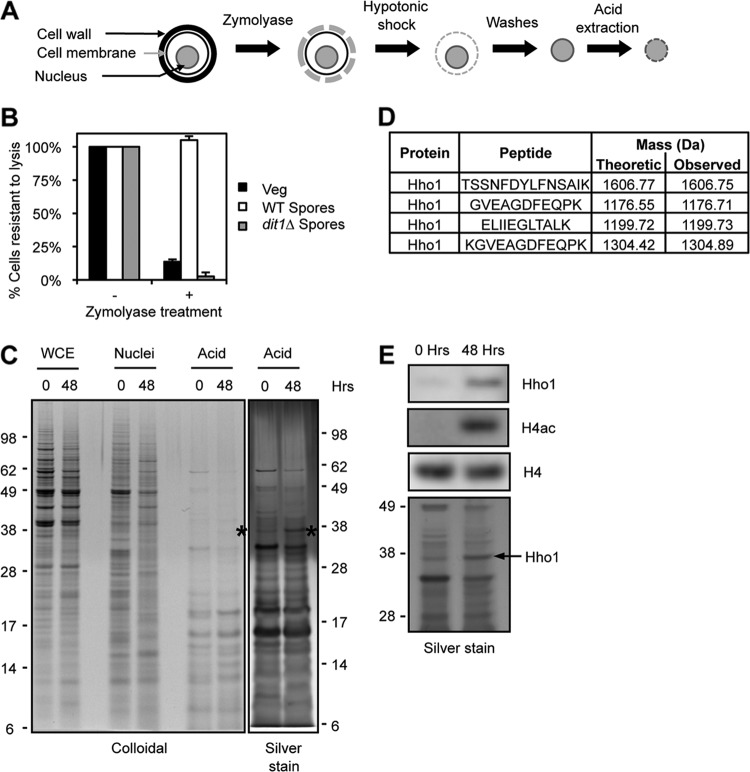
Fig 1.
Differential analysis of spore chromatin and identification of Hho1. (A) Purification scheme. The cell wall is first digested with Zymolyase. Nuclei are then released by a hypotonic shock and washed. Finally, basic proteins present in the chromatin are extracted with sulfuric acid. (B) Evaluation of the Zymolyase digestion. Cells with an intact wall are resistant to lysis in 1% SDS. Vegetative cells are sensitive to Zymolyase (black bars), but wild-type spores are resistant (white bars). Mutation of the DIT1 gene impairs the maturation of the spore wall, which can then be digested by Zymolyase (gray bars). (C) Protein profiles obtained at each purification step, stained by colloidal Coomassie blue or silver stain, in vegetatively growing cells (0 h) or in mature spores (48 h). Asterisks mark Hho1. Molecular masses are indicated on the sides of the gels (kDa). (D) Hho1 peptides identified by mass spectrometry analysis of the band indicated with asterisks in panel C. (E) Western blot analysis of acid extracts reveals that Hho1 is present at higher levels in spore chromatin (48 h) than in that from yeast in presporulation medium (0 h). H4 acetylation is detected with an antibody raised against a tetra-acetylated peptide (H4K5,8,12,16Ac). Molecular masses are indicated on the side of the silver-stained gel (kDa).