Fig 1.
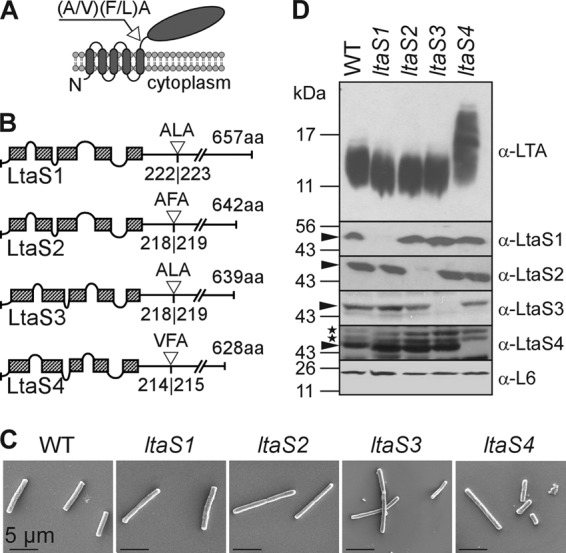
LtaS enzymes of B. anthracis. (A) Predicted topology of LtaS homologues in the plasma membrane of B. anthracis. LtaS proteins are embedded in the plasma membrane via five predicted transmembrane helices located at the N terminus of the proteins, allowing for extracellular localization of the larger catalytic domain at the C-terminal end of the protein. A single type I signal peptidase cleavage site is found following the last hydrophobic stretch, and signal peptidase cleavage effectively releases the catalytic domain (shown with arrow). (B) Linear models of LtaS enzymes of B. anthracis showing the amino acid sequences of predicted cleavage sites (shown with open triangle) and lengths of proteins. The hatched boxes represent hydrophobic regions that span the bilayer. aa, amino acids. (C) Scanning electron microscopy analysis of bacilli (wild type and ltaS1, ltaS2, ltaS3, and ltaS4 mutants) serially dehydrated and sputter coated with 80% platinum–20% palladium to 8 nm. Scale bar, 50 μm. (D) Synthesis of LTA in B. anthracis. Shown are immunoblot analyses of bacterial extracts isolated from the WT and ltaS1, ltaS2, ltaS4, and ltaS4 mutants by using antibodies specific for LTA, LtaS1, LtaS2, LtaS3, LtaS4, and ribosomal protein L6 (L6, loading control). The migration of protein size standards on SDS-PAGE gels is indicated in kilodaltons. Arrowheads point to processed LtaS proteins, and stars depict the positions of nonspecific cross-reactive immune species.
