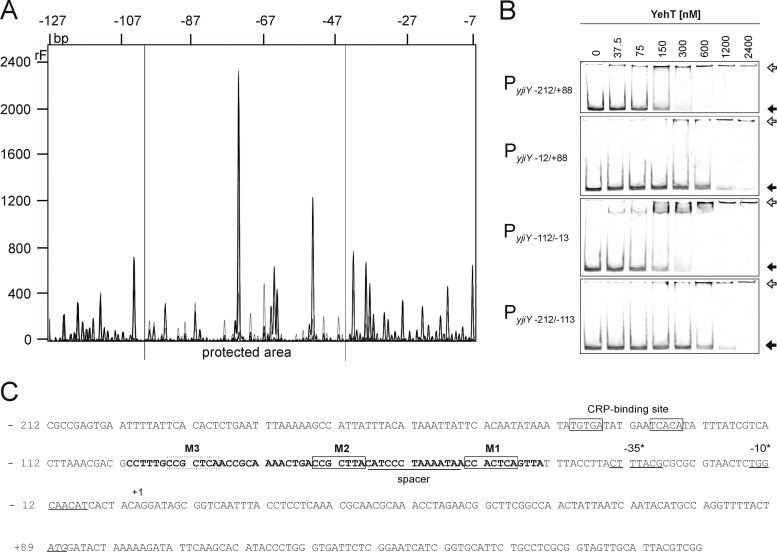
Fig 3.
Characterization of the YehT-binding site within the yjiY promoter. (A) DNase I digestion patterns were determined for DNA fragments comprising bp −212 to +88 of the yjiY promoter that had been incubated in the presence of 2.4 μM 6His-YehT (black) or BSA (gray). The DNA was labeled with fluorescein at the 5′ end of the top strand. The protected region lies within the gray horizontal lines (−101 to −44 bp). (B) EMSA of fluorescence-labeled DNA fragments comprising different segments of the yjiY promoter region following incubation with the indicated concentrations of purified 6His-YehT. The largest fragment (PyjiY−212/+88) includes the +1 transcriptional start site and the −35/−10 boxes. The positions of free DNA (filled arrows) and YehT-DNA complexes (shaded arrows) are marked. (C) Nucleotide sequence of the yjiY upstream region (positions −212 to +189). Predicted −35 and −10 promoter motifs (BProm, SoftBerry) and the transcription start site (+1) identified by 5′ RACE are indicated. The start codon is italicized and underlined, and the CRP-binding site (6) is boxed. The region protected from digestion by DNase I is marked in boldface. This 58-bp stretch includes a triple repeat of the motif CC(G/A)CT(C/T)A (M1, M2, M3) (boxed), which was identified using the MEME server (http://meme.sdsc.edu/meme/intro.html).